Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
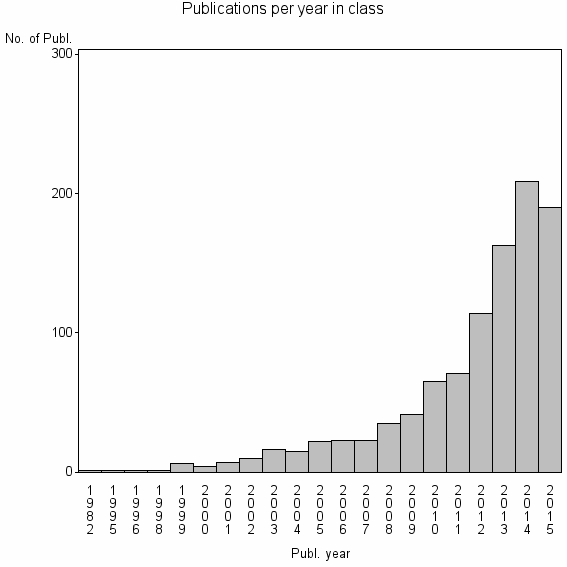
| 10217 | 1018 | 48.7 | 86% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EVOLUT CANC | Address | 9 | 36% | 2% | 20 |
| 2 | PETOS PARADOX | Author keyword | 8 | 70% | 1% | 7 |
| 3 | TRANSLAT CANC THER EUT | Address | 6 | 31% | 2% | 17 |
| 4 | CREEC | Address | 4 | 67% | 0% | 4 |
| 5 | CANC UK LUNG CANC EXCELLENCE | Address | 4 | 75% | 0% | 3 |
| 6 | CANCER GENOME | Author keyword | 4 | 27% | 1% | 13 |
| 7 | CANCER GENOMES | Author keyword | 4 | 56% | 0% | 5 |
| 8 | INTRATUMOUR HETEROGENEITY | Author keyword | 4 | 44% | 1% | 7 |
| 9 | CANC GENOME PROJECT | Address | 4 | 14% | 2% | 25 |
| 10 | CRUK LUNG CANC EXCELLENCE | Address | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PETOS PARADOX | 8 | 70% | 1% | 7 | Search PETOS+PARADOX | Search PETOS+PARADOX |
| 2 | CANCER GENOME | 4 | 27% | 1% | 13 | Search CANCER+GENOME | Search CANCER+GENOME |
| 3 | CANCER GENOMES | 4 | 56% | 0% | 5 | Search CANCER+GENOMES | Search CANCER+GENOMES |
| 4 | INTRATUMOUR HETEROGENEITY | 4 | 44% | 1% | 7 | Search INTRATUMOUR+HETEROGENEITY | Search INTRATUMOUR+HETEROGENEITY |
| 5 | ONCOGENETIC TREE | 3 | 100% | 0% | 3 | Search ONCOGENETIC+TREE | Search ONCOGENETIC+TREE |
| 6 | MUTATOR HYPOTHESIS | 3 | 60% | 0% | 3 | Search MUTATOR+HYPOTHESIS | Search MUTATOR+HYPOTHESIS |
| 7 | CANCER EVOLUTION | 2 | 24% | 1% | 9 | Search CANCER+EVOLUTION | Search CANCER+EVOLUTION |
| 8 | DRIVER MUTATIONS | 2 | 24% | 1% | 9 | Search DRIVER+MUTATIONS | Search DRIVER+MUTATIONS |
| 9 | BRANCHING TREES | 2 | 67% | 0% | 2 | Search BRANCHING+TREES | Search BRANCHING+TREES |
| 10 | CANCER HALLMARK | 2 | 67% | 0% | 2 | Search CANCER+HALLMARK | Search CANCER+HALLMARK |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | 21 BREAST CANCERS | 33 | 58% | 4% | 38 |
| 2 | MUTATIONAL EVOLUTION | 22 | 58% | 2% | 25 |
| 3 | CLONAL EVOLUTION | 19 | 17% | 10% | 100 |
| 4 | INTRATUMOR HETEROGENEITY | 14 | 25% | 5% | 51 |
| 5 | CONJUNCTIVE BAYESIAN NETWORKS | 11 | 78% | 1% | 7 |
| 6 | ONCOGENETIC TREE | 9 | 83% | 0% | 5 |
| 7 | TREE MODELS | 7 | 30% | 2% | 20 |
| 8 | CORE PATHWAYS | 7 | 39% | 1% | 13 |
| 9 | MUTAGENETIC TREES | 6 | 80% | 0% | 4 |
| 10 | MUTATED DRIVER PATHWAYS | 6 | 80% | 0% | 4 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Cancer Genome Landscapes | 2013 | 802 | 139 | 17% |
| Clonal evolution in cancer | 2012 | 429 | 89 | 38% |
| Predictive genomics: A cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data | 2015 | 4 | 25 | 52% |
| The cancer genome | 2009 | 760 | 48 | 19% |
| Exploring the Genomes of Cancer Cells: Progress and Promise | 2011 | 233 | 66 | 29% |
| Intra-tumour heterogeneity: a looking glass for cancer? | 2012 | 297 | 94 | 31% |
| The causes and consequences of genetic heterogeneity in cancer evolution | 2013 | 170 | 118 | 25% |
| Intratumor Heterogeneity: Evolution through Space and Time | 2012 | 122 | 35 | 54% |
| Cancer heterogeneity: implications for targeted therapeutics | 2013 | 68 | 44 | 36% |
| Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine | 2014 | 16 | 127 | 35% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EVOLUT CANC | 9 | 36% | 2.0% | 20 |
| 2 | TRANSLAT CANC THER EUT | 6 | 31% | 1.7% | 17 |
| 3 | CREEC | 4 | 67% | 0.4% | 4 |
| 4 | CANC UK LUNG CANC EXCELLENCE | 4 | 75% | 0.3% | 3 |
| 5 | CANC GENOME PROJECT | 4 | 14% | 2.5% | 25 |
| 6 | CRUK LUNG CANC EXCELLENCE | 3 | 100% | 0.3% | 3 |
| 7 | INCA SYNERGIE | 3 | 100% | 0.3% | 3 |
| 8 | BBS PROGRAM | 2 | 67% | 0.2% | 2 |
| 9 | LUDWIG CANC GENET THER EUT | 2 | 13% | 1.5% | 15 |
| 10 | CANC BIOL EVOLUT PROGRAM | 2 | 40% | 0.4% | 4 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000189965 | BAP1//BRCA1 ASSOCIATED PROTEIN 1//SETD2 |
| 2 | 0.0000173157 | GENOME THEORY//GENOME CHAOS//REPLICATION PATTERN |
| 3 | 0.0000114166 | GENOME ASSEMBLY//VARIANT CALLING//TARGET ENRICHMENT |
| 4 | 0.0000091842 | NSSNP//POLYPHEN//NSSNPS |
| 5 | 0.0000090518 | MVK MODEL//CLONAL GROWTH MODEL//TSCE MODEL |
| 6 | 0.0000090282 | IDH1//IDH2//ISOCITRATE DEHYDROGENASE 1 |
| 7 | 0.0000088993 | COPY NUMBER VARIATION//CCL3L1//PENNCNV |
| 8 | 0.0000082810 | INTERACTION CONDITION//TARGETED CLINICAL TRIALS//INFLUENCE CONDITION |
| 9 | 0.0000078657 | FP3//HOLLOW FIBRE ASSAY//AFFILIATED ZHUJI HOSP |
| 10 | 0.0000075542 | OLAPARIB//SYNTHETIC LETHALITY//PARP INHIBITOR |