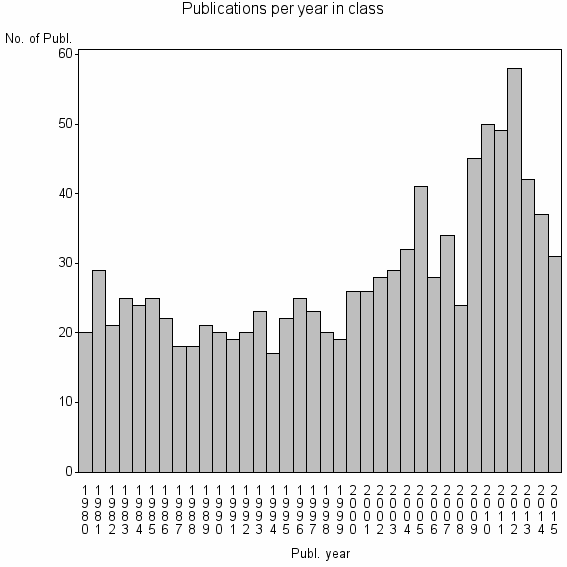
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 10279 | 1011 | 41.5 | 78% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 201 | 20439 | MELAS//MITOCHONDRIAL DNA//LEBERS HEREDITARY OPTIC NEUROPATHY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MITOCHONDRIAL NUCLEOIDS | Author keyword | 24 | 82% | 1% | 14 |
| 2 | TFAM | Author keyword | 16 | 39% | 3% | 33 |
| 3 | MITOCHONDRIAL NUCLEOID | Author keyword | 15 | 71% | 1% | 12 |
| 4 | POLRMT | Author keyword | 14 | 100% | 1% | 7 |
| 5 | ABF2 | Author keyword | 12 | 86% | 1% | 6 |
| 6 | TFB2M | Author keyword | 12 | 86% | 1% | 6 |
| 7 | MTERF | Author keyword | 10 | 73% | 1% | 8 |
| 8 | ABF2P | Author keyword | 8 | 100% | 0% | 5 |
| 9 | ATAD3 | Author keyword | 8 | 100% | 0% | 5 |
| 10 | MITOCHONDRIAL TRANSCRIPTION TERMINATION FACTOR | Author keyword | 8 | 100% | 0% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MITOCHONDRIAL NUCLEOIDS | 24 | 82% | 1% | 14 | Search MITOCHONDRIAL+NUCLEOIDS | Search MITOCHONDRIAL+NUCLEOIDS |
| 2 | TFAM | 16 | 39% | 3% | 33 | Search TFAM | Search TFAM |
| 3 | MITOCHONDRIAL NUCLEOID | 15 | 71% | 1% | 12 | Search MITOCHONDRIAL+NUCLEOID | Search MITOCHONDRIAL+NUCLEOID |
| 4 | POLRMT | 14 | 100% | 1% | 7 | Search POLRMT | Search POLRMT |
| 5 | ABF2 | 12 | 86% | 1% | 6 | Search ABF2 | Search ABF2 |
| 6 | TFB2M | 12 | 86% | 1% | 6 | Search TFB2M | Search TFB2M |
| 7 | MTERF | 10 | 73% | 1% | 8 | Search MTERF | Search MTERF |
| 8 | ABF2P | 8 | 100% | 0% | 5 | Search ABF2P | Search ABF2P |
| 9 | ATAD3 | 8 | 100% | 0% | 5 | Search ATAD3 | Search ATAD3 |
| 10 | MITOCHONDRIAL TRANSCRIPTION TERMINATION FACTOR | 8 | 100% | 0% | 5 | Search MITOCHONDRIAL+TRANSCRIPTION+TERMINATION+FACTOR | Search MITOCHONDRIAL+TRANSCRIPTION+TERMINATION+FACTOR |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MTDNA TRANSCRIPTION | 94 | 97% | 3% | 28 |
| 2 | HMG BOX PROTEIN | 40 | 64% | 4% | 39 |
| 3 | TERMINATION FACTOR MTERF | 35 | 81% | 2% | 21 |
| 4 | TRANSCRIPTION FACTOR A | 34 | 32% | 9% | 88 |
| 5 | TFAM | 33 | 83% | 2% | 19 |
| 6 | HEAVY STRAND | 30 | 68% | 3% | 27 |
| 7 | HISTONE HM | 29 | 88% | 1% | 14 |
| 8 | RNA METHYLTRANSFERASE ACTIVITY | 26 | 100% | 1% | 11 |
| 9 | LIGHT STRAND TRANSCRIPTION | 24 | 91% | 1% | 10 |
| 10 | MTDNA COPY NUMBER | 18 | 48% | 3% | 28 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number | 2012 | 54 | 104 | 62% |
| Initiation and beyond: Multiple functions of the human mitochondril transcription machinery | 2006 | 130 | 92 | 70% |
| mtDNA makes a U-turn for the mitochondrial nucleoid | 2013 | 20 | 80 | 65% |
| DNA replication and transcription in mammalian mitochondria | 2007 | 244 | 128 | 46% |
| The organization and inheritance of the mitochondrial genome | 2005 | 200 | 109 | 50% |
| Accessorizing the human mitochondrial transcription machinery | 2013 | 15 | 81 | 68% |
| Emerging functions of mammalian and plant mTERFs | 2015 | 2 | 105 | 46% |
| Functional Organization of Mammalian Mitochondrial DNA in Nucleoids: History, Recent Developments, and Future Challenges | 2010 | 61 | 109 | 43% |
| Replicating animal mitochondrial DNA | 2013 | 14 | 52 | 56% |
| Mitochondrial DNA nucleoid structure | 2012 | 41 | 85 | 39% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GEN MICROBIAL BIOCHEM | 6 | 100% | 0.4% | 4 |
| 2 | PLANT MOL BIOL BOT | 2 | 67% | 0.2% | 2 |
| 3 | MED BIOCHEM CELL BIOL | 2 | 11% | 1.9% | 19 |
| 4 | STUDI MITOCONDRI METAB ENERGET | 2 | 29% | 0.6% | 6 |
| 5 | DIPARTIMENTO BIOCHIM BIOL MOL ERNESTO QUAGLIARI | 1 | 38% | 0.3% | 3 |
| 6 | BIOQUIM B19 | 1 | 100% | 0.2% | 2 |
| 7 | SEZ BIOINFORMAT GENOM BARI | 1 | 40% | 0.2% | 2 |
| 8 | BIOL MOL ZELLBIOL PFLANZEN | 1 | 50% | 0.1% | 1 |
| 9 | MED MED REPROD BIOL | 1 | 50% | 0.1% | 1 |
| 10 | MINIST EDUC OURCE BIOL BIOTECHNOL WESTERN CH | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000288256 | DAP3//ICT1//MITOCHONDRIAL RIBOSOMAL PROTEINS |
| 2 | 0.0000163418 | H E STOICHIOMETRY//MITOCHONDRIAL PROTEIN//ALLOSTERIC ATP INHIBITION |
| 3 | 0.0000162740 | MITOCHONDRIAL DNA//MITOCHONDRIAL GENOME//MITOGENOME |
| 4 | 0.0000160065 | PODOSPORA ANSERINA//LINEAR MITOCHONDRIAL PLASMID//KALILO |
| 5 | 0.0000153581 | POLG//MNGIE//DEOXYRIBONUCLEOSIDE KINASE |
| 6 | 0.0000150940 | 4977 BP DELETION//MTDNA MUTATOR MICE//COMMON DELETION |
| 7 | 0.0000132642 | DOUBLY UNIPARENTAL INHERITANCE//PATERNAL LEAKAGE//MITOCHONDRIAL POLARITY |
| 8 | 0.0000113966 | D310//MITOCHONDRIAL MICROSATELLITE INSTABILITY//MITOCHONDRIAL HAPLOGROUPS |
| 9 | 0.0000113092 | UNITE ENDOCRINOL CELLULAIRE//DIIODOTHYRONINE//MU CRYSTALLIN |
| 10 | 0.0000095766 | DREF//INSECT BIOMED//DNA REPLICATION RELATED GENE |