Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
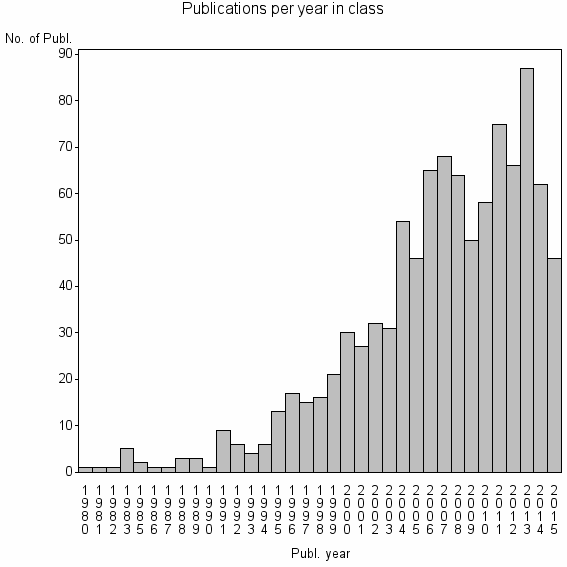
| 10565 | 987 | 35.5 | 84% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2168 | 4502 | CELL FREE PROTEIN SYNTHESIS//MEMBRANE PROTEIN//RIBOSOME DISPLAY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CELL FREE PROTEIN SYNTHESIS | Author keyword | 169 | 63% | 17% | 169 |
| 2 | RIBOSOME DISPLAY | Author keyword | 36 | 48% | 5% | 54 |
| 3 | S30 EXTRACT | Author keyword | 30 | 100% | 1% | 12 |
| 4 | MRNA DISPLAY | Author keyword | 28 | 61% | 3% | 30 |
| 5 | CELL FREE SYNTHETIC BIOLOGY | Author keyword | 21 | 90% | 1% | 9 |
| 6 | CELL FREE EXPRESSION | Author keyword | 20 | 48% | 3% | 30 |
| 7 | CELL FREE BIOLOGY | Author keyword | 18 | 83% | 1% | 10 |
| 8 | CELL FREE PROTEIN SYNTHESIS SYSTEM | Author keyword | 15 | 67% | 1% | 14 |
| 9 | IN VITRO VIRUS | Author keyword | 12 | 86% | 1% | 6 |
| 10 | CELL FREE | Author keyword | 11 | 31% | 3% | 29 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CELL FREE PROTEIN SYNTHESIS | 169 | 63% | 17% | 169 | Search CELL+FREE+PROTEIN+SYNTHESIS | Search CELL+FREE+PROTEIN+SYNTHESIS |
| 2 | RIBOSOME DISPLAY | 36 | 48% | 5% | 54 | Search RIBOSOME+DISPLAY | Search RIBOSOME+DISPLAY |
| 3 | S30 EXTRACT | 30 | 100% | 1% | 12 | Search S30+EXTRACT | Search S30+EXTRACT |
| 4 | MRNA DISPLAY | 28 | 61% | 3% | 30 | Search MRNA+DISPLAY | Search MRNA+DISPLAY |
| 5 | CELL FREE SYNTHETIC BIOLOGY | 21 | 90% | 1% | 9 | Search CELL+FREE+SYNTHETIC+BIOLOGY | Search CELL+FREE+SYNTHETIC+BIOLOGY |
| 6 | CELL FREE EXPRESSION | 20 | 48% | 3% | 30 | Search CELL+FREE+EXPRESSION | Search CELL+FREE+EXPRESSION |
| 7 | CELL FREE BIOLOGY | 18 | 83% | 1% | 10 | Search CELL+FREE+BIOLOGY | Search CELL+FREE+BIOLOGY |
| 8 | CELL FREE PROTEIN SYNTHESIS SYSTEM | 15 | 67% | 1% | 14 | Search CELL+FREE+PROTEIN+SYNTHESIS+SYSTEM | Search CELL+FREE+PROTEIN+SYNTHESIS+SYSTEM |
| 9 | IN VITRO VIRUS | 12 | 86% | 1% | 6 | Search IN+VITRO+VIRUS | Search IN+VITRO+VIRUS |
| 10 | CELL FREE | 11 | 31% | 3% | 29 | Search CELL+FREE | Search CELL+FREE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SYNTHESIS SYSTEM | 47 | 45% | 8% | 80 |
| 2 | FREE PROTEIN SYNTHESIS | 47 | 43% | 8% | 83 |
| 3 | FREE TRANSLATION | 44 | 71% | 4% | 36 |
| 4 | FREE TRANSLATION SYSTEM | 43 | 55% | 5% | 54 |
| 5 | COUPLED TRANSCRIPTION TRANSLATION | 39 | 73% | 3% | 30 |
| 6 | FREE EXPRESSION | 39 | 63% | 4% | 40 |
| 7 | FUNCTIONAL PROTEINS | 34 | 48% | 5% | 52 |
| 8 | WHEAT GERM EXTRACT | 27 | 76% | 2% | 19 |
| 9 | RIBOSOME DISPLAY | 25 | 30% | 7% | 69 |
| 10 | AMINO ACID STABILIZATION | 23 | 100% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Cell-free protein synthesis: Applications come of age | 2012 | 64 | 86 | 73% |
| Engine out of the chassis: Cell-free protein synthesis and its uses | 2014 | 9 | 68 | 49% |
| The past, present and future of cell-free protein synthesis | 2005 | 134 | 46 | 63% |
| High-throughput cell-free systems for synthesis of functionally active proteins | 2004 | 109 | 48 | 85% |
| Production of membrane proteins using cell-free expression systems | 2008 | 49 | 65 | 75% |
| Membrane protein expression: no cells required | 2009 | 52 | 49 | 45% |
| Cell-free expression as an emerging technique for the large scale production of integral membrane protein | 2006 | 53 | 45 | 58% |
| Cell-free expression systems for eukaryotic protein production | 2006 | 71 | 37 | 43% |
| In-vitro protein evolution by ribosome display and mRNA display | 2004 | 175 | 67 | 43% |
| Methods for energizing cell-free protein synthesis | 2009 | 13 | 22 | 95% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ICTAS | 9 | 29% | 2.5% | 25 |
| 2 | CELL FREE SCI TECHNOL | 5 | 14% | 3.4% | 34 |
| 3 | BIOL AGR SCI | 4 | 22% | 1.6% | 16 |
| 4 | FINE CHEM ENGN CHEM | 4 | 13% | 2.8% | 28 |
| 5 | SAITAMA IND TECHNOL | 3 | 50% | 0.5% | 5 |
| 6 | INNOVAT START UPS | 3 | 100% | 0.3% | 3 |
| 7 | JANUSYS CORP | 3 | 100% | 0.3% | 3 |
| 8 | SAITAMA SMALL ENTERPRISE PROMOT CORP | 3 | 60% | 0.3% | 3 |
| 9 | CLIN BIOTECHNOL BUSINESS UNITNAKAGYO KU | 2 | 67% | 0.2% | 2 |
| 10 | MICROBIAL METAB MOE | 2 | 67% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000153249 | PYRROLYSYL TRNA SYNTHETASE//PYRROLYSINE//GENETIC CODE EXPANSION |
| 2 | 0.0000139163 | AMPHIPOLS//UMR 7099//AMPHIPOL |
| 3 | 0.0000118836 | PROTOCELL//ELECTROFORMATION//MINIMAL CELL |
| 4 | 0.0000094276 | DIRECTED EVOLUTION//DNA SHUFFLING//EPPCR |
| 5 | 0.0000081525 | TEV PROTEASE//SOLUBILITY ENHANCER//FUSION TAG |
| 6 | 0.0000071865 | SCFV//SINGLE CHAIN FV//PHAGE DISPLAY |
| 7 | 0.0000069443 | TRIGGER FACTOR//NASCENT CHAIN//COTRANSLATIONAL FOLDING |
| 8 | 0.0000067434 | AFFIBODY MOLECULE//AFFIBODY MOLECULES//AFFIBODY |
| 9 | 0.0000058629 | ANTIBODY MICROARRAYS//PROTEIN MICROARRAY//PROTEIN MICROARRAYS |
| 10 | 0.0000048771 | T VECTOR//HIGH THROUGHPUT CLONING//TA CLONING |