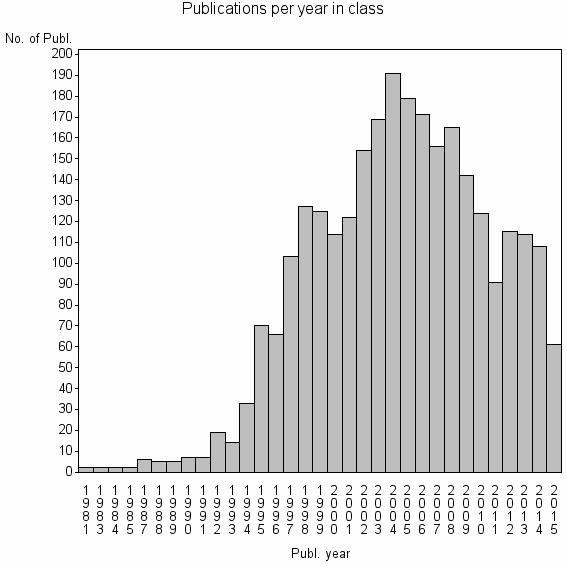
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 1071 | 2771 | 49.5 | 86% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN FOLDING | Author keyword | 70 | 11% | 22% | 615 |
| 2 | PHI VALUE | Author keyword | 65 | 77% | 2% | 44 |
| 3 | CONTACT ORDER | Author keyword | 60 | 80% | 1% | 37 |
| 4 | PHI VALUE ANALYSIS | Author keyword | 42 | 69% | 1% | 36 |
| 5 | FOLDING RATE | Author keyword | 39 | 73% | 1% | 30 |
| 6 | FOLDING KINETICS | Author keyword | 38 | 41% | 3% | 71 |
| 7 | FOLDING NUCLEUS | Author keyword | 35 | 67% | 1% | 32 |
| 8 | NUCLEATION CONDENSATION | Author keyword | 33 | 100% | 0% | 13 |
| 9 | PHI VALUES | Author keyword | 32 | 85% | 1% | 17 |
| 10 | TRANSITION STATE ENSEMBLE | Author keyword | 31 | 76% | 1% | 22 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN FOLDING | 70 | 11% | 22% | 615 | Search PROTEIN+FOLDING | Search PROTEIN+FOLDING |
| 2 | PHI VALUE | 65 | 77% | 2% | 44 | Search PHI+VALUE | Search PHI+VALUE |
| 3 | CONTACT ORDER | 60 | 80% | 1% | 37 | Search CONTACT+ORDER | Search CONTACT+ORDER |
| 4 | PHI VALUE ANALYSIS | 42 | 69% | 1% | 36 | Search PHI+VALUE+ANALYSIS | Search PHI+VALUE+ANALYSIS |
| 5 | FOLDING RATE | 39 | 73% | 1% | 30 | Search FOLDING+RATE | Search FOLDING+RATE |
| 6 | FOLDING KINETICS | 38 | 41% | 3% | 71 | Search FOLDING+KINETICS | Search FOLDING+KINETICS |
| 7 | FOLDING NUCLEUS | 35 | 67% | 1% | 32 | Search FOLDING+NUCLEUS | Search FOLDING+NUCLEUS |
| 8 | NUCLEATION CONDENSATION | 33 | 100% | 0% | 13 | Search NUCLEATION+CONDENSATION | Search NUCLEATION+CONDENSATION |
| 9 | PHI VALUES | 32 | 85% | 1% | 17 | Search PHI+VALUES | Search PHI+VALUES |
| 10 | TRANSITION STATE ENSEMBLE | 31 | 76% | 1% | 22 | Search TRANSITION+STATE+ENSEMBLE | Search TRANSITION+STATE+ENSEMBLE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CONTACT ORDER | 341 | 73% | 9% | 260 |
| 2 | CHYMOTRYPSIN INHIBITOR 2 | 296 | 63% | 11% | 299 |
| 3 | SINGLE DOMAIN PROTEINS | 238 | 76% | 6% | 166 |
| 4 | FUNNELS | 175 | 61% | 7% | 184 |
| 5 | TRANSITION STATE PLACEMENT | 114 | 95% | 1% | 38 |
| 6 | TRANSITION STATE | 111 | 15% | 25% | 697 |
| 7 | 2 STATE PROTEINS | 111 | 76% | 3% | 77 |
| 8 | NUCLEATION CONDENSATION MECHANISM | 95 | 66% | 3% | 88 |
| 9 | TRANSITION STATE ENSEMBLE | 95 | 68% | 3% | 83 |
| 10 | 2 STATE TRANSITION | 93 | 88% | 2% | 44 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FOLDING & DESIGN | 5 | 16% | 1% | 29 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The protein folding problem | 2008 | 348 | 213 | 49% |
| The protein folding 'speed limit' | 2004 | 475 | 137 | 72% |
| From Levinthal to pathways to funnels | 1997 | 1345 | 74 | 59% |
| Protein folding thermodynamics and dynamics: Where physics, chemistry, and biology meet | 2006 | 195 | 238 | 62% |
| Coarse-grained models of protein folding: toy models or predictive tools? | 2008 | 133 | 55 | 55% |
| Intermediates: ubiquitous species on folding energy landscapes? | 2007 | 106 | 65 | 71% |
| Is there a unifying mechanism for protein folding? | 2003 | 265 | 44 | 70% |
| Insights from Coarse-Grained Go Models for Protein Folding and Dynamics | 2009 | 90 | 150 | 52% |
| The experimental survey of protein-folding energy landscapes | 2005 | 123 | 158 | 84% |
| The Protein-Folding Problem, 50 Years On | 2012 | 168 | 61 | 20% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | THEORET BIOL PHYS | 9 | 10% | 2.9% | 80 |
| 2 | CHIM BIOPHYS | 6 | 16% | 1.3% | 35 |
| 3 | PROGRAM BIOCHEM STRUCT BIOL | 6 | 21% | 0.9% | 26 |
| 4 | MRC PROT ENGN | 5 | 42% | 0.4% | 10 |
| 5 | MED COUNCIL PROT ENGN | 4 | 75% | 0.1% | 3 |
| 6 | QUANTITAT BIOSCI BERKELEY | 4 | 75% | 0.1% | 3 |
| 7 | BIOMOL STRUCT DESIGN PROGRAM | 4 | 22% | 0.6% | 16 |
| 8 | GLOBAL STRATEG ANALYT UNIT | 3 | 100% | 0.1% | 3 |
| 9 | BIOL TEOR | 3 | 37% | 0.3% | 7 |
| 10 | CAMBRIDGE PROT ENGN | 3 | 23% | 0.4% | 12 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000224346 | HP MODEL//STATISTICAL POTENTIALS//PROTEIN STRUCTURE PREDICTION |
| 2 | 0.0000166569 | BETA HAIRPIN//SIDE CHAIN INTERACTIONS//TRP CAGE |
| 3 | 0.0000154467 | MOLTEN GLOBULE//ALPHA LACTALBUMIN//APOMYOGLOBIN |
| 4 | 0.0000121293 | MARKOV STATE MODELS//TRANSITION PATH SAMPLING//TRANSITION PATH THEORY |
| 5 | 0.0000119174 | POLYPROLINE II//RANDOM COIL//COIL LIBRARY |
| 6 | 0.0000106415 | BASE PAIR KINETICS//GNRA TETRALOOPS//RAAA HAIRPIN MOTIF |
| 7 | 0.0000103723 | HYDROGEN EXCHANGE//HYDROGEN DEUTERIUM EXCHANGE//AMIDE HYDROGEN EXCHANGE |
| 8 | 0.0000103349 | BAKER CHEM CHEM BIOL//REPLICA EXCHANGE METHOD//GENERALIZED ENSEMBLE ALGORITHM |
| 9 | 0.0000097467 | DEHYDRON//KINASE TARGETING//ONCOKINOME |
| 10 | 0.0000095444 | STAPHYLOCOCCAL NUCLEASE//THERMODYNAMIC MOLECULAR SWITCH//PROTEIN THERMOSTABILITY |