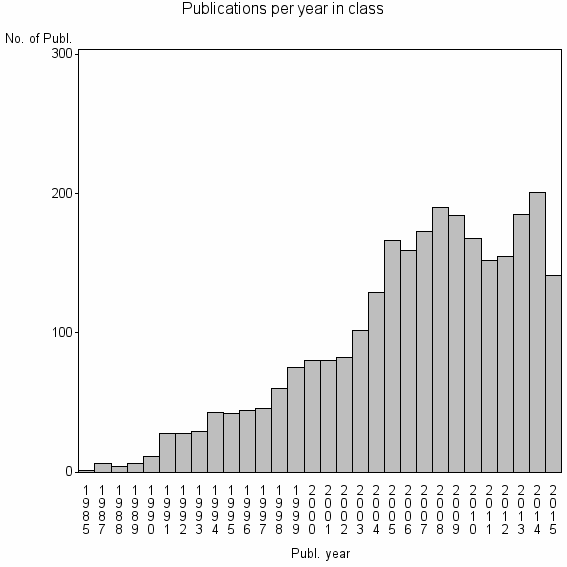
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 1072 | 2770 | 51.9 | 93% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 195 | 20594 | PRE MRNA SPLICING//RNA LOCALIZATION//SPLICEOSOME |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SR PROTEINS | Author keyword | 93 | 60% | 4% | 101 |
| 2 | SR PROTEIN | Author keyword | 63 | 71% | 2% | 50 |
| 3 | ALTERNATIVE SPLICING | Author keyword | 60 | 11% | 18% | 502 |
| 4 | SRPK1 | Author keyword | 33 | 83% | 1% | 19 |
| 5 | POLYPYRIMIDINE TRACT BINDING PROTEIN | Author keyword | 28 | 56% | 1% | 34 |
| 6 | SPLICING FACTORS | Author keyword | 24 | 35% | 2% | 57 |
| 7 | SPLICING REGULATORY ELEMENTS | Author keyword | 21 | 90% | 0% | 9 |
| 8 | RNA RNP GRP | Address | 17 | 100% | 0% | 8 |
| 9 | RS DOMAIN | Author keyword | 16 | 64% | 1% | 16 |
| 10 | SF2 ASF | Author keyword | 16 | 65% | 1% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SR PROTEINS | 93 | 60% | 4% | 101 | Search SR+PROTEINS | Search SR+PROTEINS |
| 2 | SR PROTEIN | 63 | 71% | 2% | 50 | Search SR+PROTEIN | Search SR+PROTEIN |
| 3 | ALTERNATIVE SPLICING | 60 | 11% | 18% | 502 | Search ALTERNATIVE+SPLICING | Search ALTERNATIVE+SPLICING |
| 4 | SRPK1 | 33 | 83% | 1% | 19 | Search SRPK1 | Search SRPK1 |
| 5 | POLYPYRIMIDINE TRACT BINDING PROTEIN | 28 | 56% | 1% | 34 | Search POLYPYRIMIDINE+TRACT+BINDING+PROTEIN | Search POLYPYRIMIDINE+TRACT+BINDING+PROTEIN |
| 6 | SPLICING FACTORS | 24 | 35% | 2% | 57 | Search SPLICING+FACTORS | Search SPLICING+FACTORS |
| 7 | SPLICING REGULATORY ELEMENTS | 21 | 90% | 0% | 9 | Search SPLICING+REGULATORY+ELEMENTS | Search SPLICING+REGULATORY+ELEMENTS |
| 8 | RS DOMAIN | 16 | 64% | 1% | 16 | Search RS+DOMAIN | Search RS+DOMAIN |
| 9 | SF2 ASF | 16 | 65% | 1% | 15 | Search SF2+ASF | Search SF2+ASF |
| 10 | SPLICING ENHANCERS | 15 | 88% | 0% | 7 | Search SPLICING+ENHANCERS | Search SPLICING+ENHANCERS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SR PROTEINS | 413 | 53% | 20% | 551 |
| 2 | PRE MESSENGER RNA | 222 | 20% | 35% | 966 |
| 3 | RS DOMAIN | 94 | 76% | 2% | 65 |
| 4 | TRACT BINDING PROTEIN | 85 | 31% | 8% | 232 |
| 5 | FACTOR ASF SF2 | 71 | 75% | 2% | 51 |
| 6 | SERINE RICH | 64 | 68% | 2% | 56 |
| 7 | DEFAULT EXON | 53 | 95% | 1% | 18 |
| 8 | ASF SF2 | 50 | 76% | 1% | 35 |
| 9 | FACTORS ASF SF2 | 49 | 94% | 1% | 17 |
| 10 | T ALTERNATIVE EXON | 47 | 81% | 1% | 29 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Expansion of the eukaryotic proteome by alternative splicing | 2010 | 501 | 81 | 48% |
| The SR protein family of splicing factors: master regulators of gene expression | 2009 | 373 | 206 | 67% |
| Alternative splicing: New insights from global analyses | 2006 | 527 | 76 | 71% |
| Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches | 2009 | 366 | 160 | 61% |
| Understanding alternative splicing: Towards a cellular code | 2005 | 588 | 134 | 79% |
| Context-dependent control of alternative splicing by RNA-binding proteins | 2014 | 20 | 170 | 75% |
| Splicing regulation: From a parts list of regulatory elements to an integrated splicing code | 2008 | 319 | 140 | 71% |
| Alternative splicing: a pivotal step between eukaryotic transcription and translation | 2013 | 136 | 131 | 34% |
| Mechanisms of alternative pre-messenger RNA splicing | 2003 | 1260 | 352 | 49% |
| Functional consequences of developmentally regulated alternative splicing | 2011 | 153 | 160 | 49% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RNA RNP GRP | 17 | 100% | 0.3% | 8 |
| 2 | GENOM PHYSIOL | 7 | 56% | 0.3% | 9 |
| 3 | PROGRAM MOL PLANT BIOL | 7 | 32% | 0.6% | 17 |
| 4 | EQUIPE BASES MOL PROG S CANC POUMON 2 | 6 | 71% | 0.2% | 5 |
| 5 | PROGRAM MOL CANC BIOL | 4 | 47% | 0.3% | 7 |
| 6 | CORE MOL PHARMACOL | 4 | 75% | 0.1% | 3 |
| 7 | HUGHES UNDER | 4 | 75% | 0.1% | 3 |
| 8 | AGR BIG DATA | 3 | 57% | 0.1% | 4 |
| 9 | HAVANA GRP | 3 | 100% | 0.1% | 3 |
| 10 | MICROBIOL INFECTIOL | 3 | 11% | 1.0% | 27 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000166005 | SPLICEOSOME//PRE MRNA SPLICING//PRP24 |
| 2 | 0.0000123015 | HNRNP B1//HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEINS HNRNPS//RNP CONCENSUS RNA BINDING DOMAIN |
| 3 | 0.0000096590 | UPF1//NMD//NONSENSE MEDIATED MRNA DECAY |
| 4 | 0.0000091581 | P54NRB//POLYPYRIMIDINE TRACT BINDING PROTEIN ASSOCIATED SPLICING FACTOR//MATRIN 3 |
| 5 | 0.0000090391 | REV//RRE//REV PROTEIN |
| 6 | 0.0000086201 | HERBOXIDIENE//FR901464//PLADIENOLIDE |
| 7 | 0.0000083069 | INTRON GAIN//INTRON LOSS//INTRON EVOLUTION |
| 8 | 0.0000075452 | RNA SEQ//DE NOVO ASSEMBLY//TRANSCRIPTOME ASSEMBLY |
| 9 | 0.0000072084 | EXON SKIPPING//DMD GENET THER Y GRP//MC LOCKWOOD MUSCULAR DYSTROPHY |
| 10 | 0.0000071741 | INTERACTION PROPENSITY//RNA BINDING RESIDUES//COMPUTAT INTELLIGENCE LEARNING DISCOVERY |