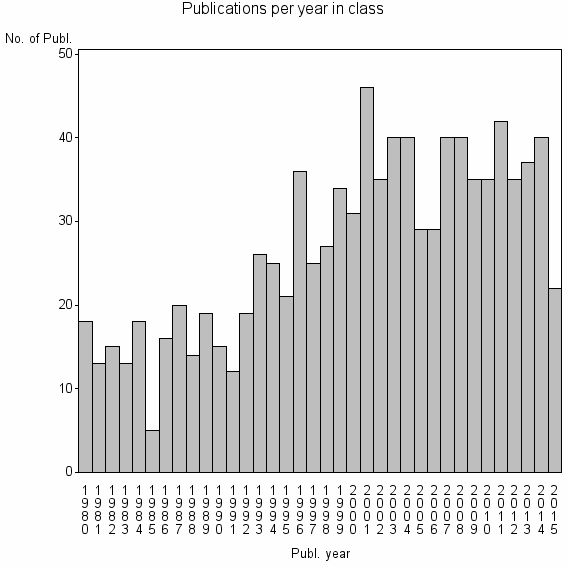
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 10781 | 967 | 39.6 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 623 | 13179 | BASE EXCISION REPAIR//DNA GLYCOSYLASE//URACIL DNA GLYCOSYLASE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | URACIL DNA GLYCOSYLASE | Author keyword | 92 | 63% | 10% | 92 |
| 2 | DUTPASE | Author keyword | 79 | 71% | 7% | 64 |
| 3 | DUTP PYROPHOSPHATASE | Author keyword | 18 | 89% | 1% | 8 |
| 4 | MBD4 | Author keyword | 13 | 55% | 2% | 16 |
| 5 | DUTP | Author keyword | 7 | 57% | 1% | 8 |
| 6 | URACIL DNA GLYCOSYLASE INHIBITOR | Author keyword | 6 | 100% | 0% | 4 |
| 7 | UNG | Author keyword | 5 | 37% | 1% | 10 |
| 8 | UDG | Author keyword | 4 | 42% | 1% | 8 |
| 9 | URACIL IN DNA | Author keyword | 4 | 75% | 0% | 3 |
| 10 | THYMINE DNA GLYCOSYLASE | Author keyword | 4 | 36% | 1% | 8 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | URACIL DNA GLYCOSYLASE | 92 | 63% | 10% | 92 | Search URACIL+DNA+GLYCOSYLASE | Search URACIL+DNA+GLYCOSYLASE |
| 2 | DUTPASE | 79 | 71% | 7% | 64 | Search DUTPASE | Search DUTPASE |
| 3 | DUTP PYROPHOSPHATASE | 18 | 89% | 1% | 8 | Search DUTP+PYROPHOSPHATASE | Search DUTP+PYROPHOSPHATASE |
| 4 | MBD4 | 13 | 55% | 2% | 16 | Search MBD4 | Search MBD4 |
| 5 | DUTP | 7 | 57% | 1% | 8 | Search DUTP | Search DUTP |
| 6 | URACIL DNA GLYCOSYLASE INHIBITOR | 6 | 100% | 0% | 4 | Search URACIL+DNA+GLYCOSYLASE+INHIBITOR | Search URACIL+DNA+GLYCOSYLASE+INHIBITOR |
| 7 | UNG | 5 | 37% | 1% | 10 | Search UNG | Search UNG |
| 8 | UDG | 4 | 42% | 1% | 8 | Search UDG | Search UDG |
| 9 | URACIL IN DNA | 4 | 75% | 0% | 3 | Search URACIL+IN+DNA | Search URACIL+IN+DNA |
| 10 | THYMINE DNA GLYCOSYLASE | 4 | 36% | 1% | 8 | Search THYMINE+DNA+GLYCOSYLASE | Search THYMINE+DNA+GLYCOSYLASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DEOXYURIDINE TRIPHOSPHATE NUCLEOTIDOHYDROLASE | 99 | 87% | 5% | 48 |
| 2 | DEOXYURIDINE TRIPHOSPHATASE | 67 | 70% | 6% | 56 |
| 3 | NUCLEOTIDOHYDROLASE | 57 | 95% | 2% | 19 |
| 4 | ESCHERICHIA COLI DUTPASE | 56 | 100% | 2% | 19 |
| 5 | UNG GENE | 40 | 70% | 3% | 33 |
| 6 | MED1 MBD4 | 25 | 73% | 2% | 19 |
| 7 | BACTERIOPHAGE PBS2 | 23 | 79% | 2% | 15 |
| 8 | DUTP PYROPHOSPHATASE | 23 | 79% | 2% | 15 |
| 9 | URACIL INCORPORATION | 19 | 68% | 2% | 17 |
| 10 | DUTPASE | 16 | 48% | 2% | 24 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Keeping Uracil Out of DNA: Physiological Role, Structure and Catalytic Mechanism of dUTPases | 2009 | 48 | 56 | 89% |
| Uracil-DNA glycosylases-Structural and functional perspectives on an essential family of DNA repair enzymes | 2014 | 6 | 74 | 70% |
| MBD4 and TDG: Multifaceted DNA glycosylases with ever expanding biological roles | 2013 | 17 | 109 | 42% |
| Participation of DNA repair in the response to 5-fluorouracil | 2009 | 69 | 87 | 41% |
| Uracil in DNA and its processing by different DNA glycosylases | 2009 | 36 | 32 | 66% |
| Uracil in DNA - occurrence, consequences and repair | 2002 | 197 | 100 | 56% |
| dUTPases, the unexplored family of signalling molecules | 2013 | 6 | 41 | 63% |
| The enigmatic thymine DNA glycosylase | 2007 | 98 | 103 | 33% |
| Preventive DNA repair by sanitizing the cellular (deoxy) nucleoside triphosphate pool | 2014 | 3 | 111 | 47% |
| Genomic uracil and human disease | 2006 | 27 | 32 | 69% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENOME METAB REPAIR | 3 | 100% | 0.3% | 3 |
| 2 | SOUTH CAROLINA EXPT STN | 2 | 67% | 0.2% | 2 |
| 3 | UMR 8113GRP REPARAT ADN | 2 | 67% | 0.2% | 2 |
| 4 | UNIGEN MOLEC BIOL | 2 | 67% | 0.2% | 2 |
| 5 | DOCTORAL MULTIDISCIPLINARY MED SCI | 1 | 50% | 0.2% | 2 |
| 6 | GENOME METAB | 1 | 100% | 0.2% | 2 |
| 7 | UPJOHN CLIN PHARMACOL 4701 | 1 | 50% | 0.2% | 2 |
| 8 | U372 | 1 | 16% | 0.6% | 6 |
| 9 | LBPA ENS CACHAN | 1 | 40% | 0.2% | 2 |
| 10 | PROT MODELING GRP | 1 | 16% | 0.5% | 5 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000203765 | ENDONUCLEASE V//S CAROLINA EXPT STN//OXANINE |
| 2 | 0.0000203691 | DNA GLYCOSYLASE//8 OXOGUANINE//BASE EXCISION REPAIR |
| 3 | 0.0000161885 | APE1//AP ENDONUCLEASE//APE1 REF 1 |
| 4 | 0.0000117731 | DNTP IMBALANCE//BLC 2//MICROBIAL TEST SYSTEMS |
| 5 | 0.0000078735 | 5 HYDROXYMETHYLCYTOSINE//5HMC//5 CARBOXYLCYTOSINE |
| 6 | 0.0000059967 | CYTIDINE DEAMINASE//ANTIBIOTIC FUNGICIDE//BLASTICIDIN S DEAMINASE |
| 7 | 0.0000059117 | BACTERIOPHAGE T5//HOST PHAGE INTERACTION//LYTIC CONVERSION |
| 8 | 0.0000053210 | MUTS//MISMATCH REPAIR//DNA MISMATCH REPAIR |
| 9 | 0.0000051891 | URIDINE PHOSPHORYLASE//5 PHENYLTHIOACYCLOURIDINE//ALPHA D RIBOSE 1 PHOSPHATE |
| 10 | 0.0000045501 | VPR//VPX//HIV 1 VPR |