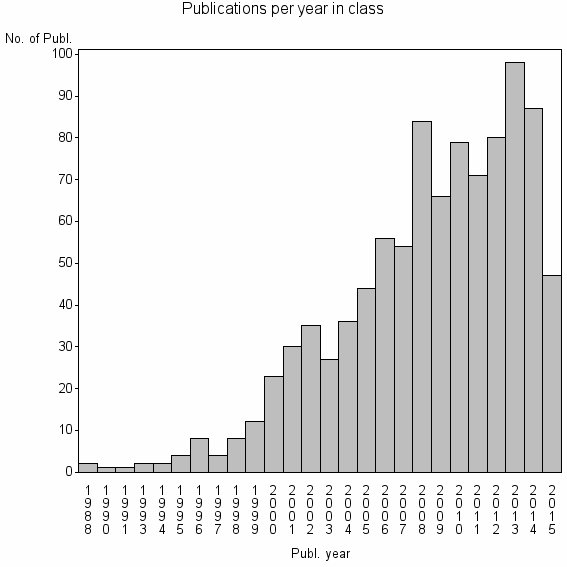
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 10861 | 961 | 53.8 | 95% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 195 | 20594 | PRE MRNA SPLICING//RNA LOCALIZATION//SPLICEOSOME |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MRNP BIOGENESIS METAB | Address | 26 | 49% | 4% | 38 |
| 2 | DIS3 | Author keyword | 17 | 72% | 1% | 13 |
| 3 | RRP6 | Author keyword | 17 | 75% | 1% | 12 |
| 4 | NUCLEAR EXOSOME | Author keyword | 14 | 100% | 1% | 7 |
| 5 | THO COMPLEX | Author keyword | 13 | 69% | 1% | 11 |
| 6 | MRNA EXPORT | Author keyword | 13 | 24% | 5% | 45 |
| 7 | MRNP BIOGENESIS | Author keyword | 12 | 86% | 1% | 6 |
| 8 | ANDALUZ BIOL MOL MED REGENERAT CABIMER | Address | 11 | 35% | 3% | 26 |
| 9 | R LOOPS | Author keyword | 11 | 54% | 1% | 14 |
| 10 | SUB2 | Author keyword | 11 | 78% | 1% | 7 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DIS3 | 17 | 72% | 1% | 13 | Search DIS3 | Search DIS3 |
| 2 | RRP6 | 17 | 75% | 1% | 12 | Search RRP6 | Search RRP6 |
| 3 | NUCLEAR EXOSOME | 14 | 100% | 1% | 7 | Search NUCLEAR+EXOSOME | Search NUCLEAR+EXOSOME |
| 4 | THO COMPLEX | 13 | 69% | 1% | 11 | Search THO+COMPLEX | Search THO+COMPLEX |
| 5 | MRNA EXPORT | 13 | 24% | 5% | 45 | Search MRNA+EXPORT | Search MRNA+EXPORT |
| 6 | MRNP BIOGENESIS | 12 | 86% | 1% | 6 | Search MRNP+BIOGENESIS | Search MRNP+BIOGENESIS |
| 7 | R LOOPS | 11 | 54% | 1% | 14 | Search R+LOOPS | Search R+LOOPS |
| 8 | SUB2 | 11 | 78% | 1% | 7 | Search SUB2 | Search SUB2 |
| 9 | TREX COMPLEX | 11 | 78% | 1% | 7 | Search TREX+COMPLEX | Search TREX+COMPLEX |
| 10 | EXOSOME | 10 | 14% | 7% | 67 | Search EXOSOME | Search EXOSOME |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | YEAST EXOSOME | 132 | 79% | 9% | 84 |
| 2 | HUMAN PM SCL | 99 | 80% | 6% | 61 |
| 3 | CRYPTIC UNSTABLE TRANSCRIPTS | 76 | 68% | 7% | 67 |
| 4 | BINDING PROTEINS NRD1 | 65 | 85% | 4% | 34 |
| 5 | THO COMPLEX | 65 | 85% | 4% | 34 |
| 6 | CORE EXOSOME | 54 | 83% | 3% | 30 |
| 7 | POLYA POLYMERASE | 48 | 26% | 17% | 159 |
| 8 | NUCLEAR EXOSOME | 46 | 75% | 3% | 33 |
| 9 | TREX COMPLEX | 45 | 68% | 4% | 39 |
| 10 | ARCHAEAL EXOSOME | 39 | 73% | 3% | 30 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| RNA decay machines: The exosome | 2013 | 38 | 97 | 74% |
| R Loops: From Transcription Byproducts to Threats to Genome Stability | 2012 | 145 | 79 | 35% |
| RNA-quality control by the exosome | 2006 | 339 | 95 | 59% |
| The exosome: a multipurpose RNA-decay machine | 2008 | 118 | 68 | 75% |
| Threading the barrel of the RNA exosome | 2013 | 22 | 80 | 86% |
| The interface between transcription and mRNP export: From THO to THSC/TREX-2 | 2010 | 36 | 73 | 79% |
| Transcription termination and the control of the transcriptome: why, where and how to stop | 2015 | 1 | 126 | 51% |
| The exosome and RNA quality control in the nucleus | 2007 | 72 | 51 | 90% |
| A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression | 2014 | 18 | 114 | 33% |
| The Many Pathways of RNA Degradation | 2009 | 304 | 110 | 27% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MRNP BIOGENESIS METAB | 26 | 49% | 4.0% | 38 |
| 2 | ANDALUZ BIOL MOL MED REGENERAT CABIMER | 11 | 35% | 2.7% | 26 |
| 3 | LEA NUCL RNA METAB | 6 | 80% | 0.4% | 4 |
| 4 | UNITE GENET INTERACT MACROMOL | 3 | 32% | 0.9% | 9 |
| 5 | GENE EXP S COUPLED RNA TRANSPORT | 2 | 67% | 0.2% | 2 |
| 6 | RNA BIOL GENOME MED | 2 | 67% | 0.2% | 2 |
| 7 | BIOMOL NETWORKS S | 1 | 31% | 0.4% | 4 |
| 8 | ANDALUZ BIOL MOL MED REGENERATIVA CABIMER | 1 | 50% | 0.2% | 2 |
| 9 | CELL BIOL FRONTIERS GENET | 1 | 100% | 0.2% | 2 |
| 10 | CABIMER | 1 | 17% | 0.5% | 5 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000164762 | DECAPPING//P BODIES//CCR4 NOT |
| 2 | 0.0000162878 | POLYADENYLATION//ALTERNATIVE POLYADENYLATION//POLYA POLYMERASE |
| 3 | 0.0000159875 | P TEFB//CDK9//HEXIM1 |
| 4 | 0.0000145177 | UPF1//NMD//NONSENSE MEDIATED MRNA DECAY |
| 5 | 0.0000123735 | SNORNA//SMALL NUCLEOLAR RNAS//PRE RRNA PROCESSING |
| 6 | 0.0000120862 | NUCLEAR PORE COMPLEX//NUCLEOPORIN//NUCLEAR TRANSPORT |
| 7 | 0.0000119420 | RNASE E//RNASE G//POLYNUCLEOTIDE PHOSPHORYLASE |
| 8 | 0.0000077689 | LIN28//LIN28B//LIN28A |
| 9 | 0.0000075651 | RNA HELICASE//DEAD BOX//DEAD BOX PROTEIN |
| 10 | 0.0000074803 | TRNA NUCLEOTIDYLTRANSFERASE//CCA ADDING ENZYME//TRNAHIS GUANYLYLTRANSFERASE |