Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
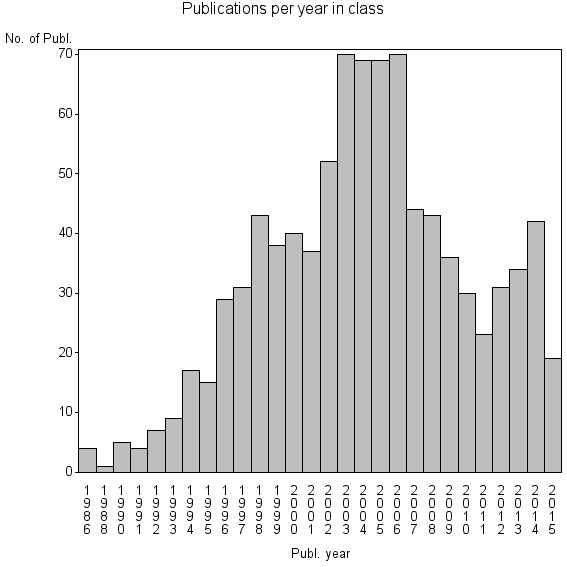
| 11477 | 912 | 30.3 | 75% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | GENE PREDICTION | Author keyword | 16 | 33% | 4% | 40 |
| 2 | GENE FINDING | Author keyword | 6 | 28% | 2% | 18 |
| 3 | GENE RECOGNITION | Author keyword | 5 | 45% | 1% | 9 |
| 4 | RE ANNOTATION | Author keyword | 3 | 50% | 1% | 5 |
| 5 | SHANDONG PROV BIOPHYS FUNCT MACROMOL | Address | 3 | 100% | 0% | 3 |
| 6 | EXON PREDICTION | Author keyword | 3 | 40% | 1% | 6 |
| 7 | SPLICED ALIGNMENT | Author keyword | 3 | 60% | 0% | 3 |
| 8 | SPLICE SITE PREDICTION | Author keyword | 3 | 35% | 1% | 6 |
| 9 | SPLICE SITES | Author keyword | 2 | 20% | 1% | 10 |
| 10 | TRANSLATION INITIATION SITE | Author keyword | 2 | 36% | 1% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENE PREDICTION | 16 | 33% | 4% | 40 | Search GENE+PREDICTION | Search GENE+PREDICTION |
| 2 | GENE FINDING | 6 | 28% | 2% | 18 | Search GENE+FINDING | Search GENE+FINDING |
| 3 | GENE RECOGNITION | 5 | 45% | 1% | 9 | Search GENE+RECOGNITION | Search GENE+RECOGNITION |
| 4 | RE ANNOTATION | 3 | 50% | 1% | 5 | Search RE+ANNOTATION | Search RE+ANNOTATION |
| 5 | EXON PREDICTION | 3 | 40% | 1% | 6 | Search EXON+PREDICTION | Search EXON+PREDICTION |
| 6 | SPLICED ALIGNMENT | 3 | 60% | 0% | 3 | Search SPLICED+ALIGNMENT | Search SPLICED+ALIGNMENT |
| 7 | SPLICE SITE PREDICTION | 3 | 35% | 1% | 6 | Search SPLICE+SITE+PREDICTION | Search SPLICE+SITE+PREDICTION |
| 8 | SPLICE SITES | 2 | 20% | 1% | 10 | Search SPLICE+SITES | Search SPLICE+SITES |
| 9 | TRANSLATION INITIATION SITE | 2 | 36% | 1% | 5 | Search TRANSLATION+INITIATION+SITE | Search TRANSLATION+INITIATION+SITE |
| 10 | COMPUTATIONAL GENE FINDING | 2 | 67% | 0% | 2 | Search COMPUTATIONAL+GENE+FINDING | Search COMPUTATIONAL+GENE+FINDING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SELF TRAINING METHOD | 34 | 93% | 1% | 13 |
| 2 | PROTEIN CODING REGIONS | 30 | 33% | 8% | 76 |
| 3 | FINDING PROGRAMS | 15 | 73% | 1% | 11 |
| 4 | GENE STRUCTURE PREDICTION | 13 | 53% | 2% | 17 |
| 5 | SPLICED ALIGNMENT | 12 | 54% | 2% | 15 |
| 6 | GENE PREDICTION | 10 | 26% | 4% | 34 |
| 7 | GENE RECOGNITION | 9 | 48% | 2% | 14 |
| 8 | PROTEIN ALIGNMENT | 8 | 100% | 1% | 5 |
| 9 | MICROBIAL GENOMES | 6 | 12% | 5% | 42 |
| 10 | GENE IDENTIFICATION | 6 | 29% | 2% | 16 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| A beginner's guide to eukaryotic genome annotation | 2012 | 82 | 105 | 32% |
| Current methods of gene prediction, their strengths and weaknesses | 2002 | 203 | 133 | 59% |
| Computational prediction of eukaryotic protein-coding genes | 2002 | 117 | 67 | 51% |
| Steady progress and recent breakthroughs in the accuracy of automated genome annotation | 2008 | 73 | 52 | 67% |
| Using comparative genome analysis to identify problems in annotated microbial genomes | 2010 | 39 | 45 | 40% |
| Between a chicken and a grape: estimating the number of human genes | 2010 | 38 | 35 | 29% |
| Finding the genes in genomic DNA | 1998 | 283 | 41 | 46% |
| EGASP: the human ENCODE genome annotation assessment project | 2006 | 49 | 57 | 44% |
| Recent advances in gene structure prediction | 2004 | 60 | 46 | 54% |
| Annotating the genome of Medicago truncatula | 2006 | 27 | 15 | 53% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SHANDONG PROV BIOPHYS FUNCT MACROMOL | 3 | 100% | 0.3% | 3 |
| 2 | ASSOCIE INRA FRANCE | 2 | 67% | 0.2% | 2 |
| 3 | SOFTWARE UNITS LAYOUT | 2 | 67% | 0.2% | 2 |
| 4 | INRA FRANCE | 2 | 50% | 0.3% | 3 |
| 5 | 659 | 1 | 100% | 0.2% | 2 |
| 6 | REGULACIO GENOM | 1 | 15% | 0.8% | 7 |
| 7 | GRP PRACT COMP SCI | 1 | 40% | 0.2% | 2 |
| 8 | ABT BIOINFORMAT | 1 | 11% | 0.8% | 7 |
| 9 | GRP RECERCA INFORMAT MED | 1 | 33% | 0.2% | 2 |
| 10 | BEREICH MOL BIOL INFORMAT | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000158029 | EMBL OUTSTN//PROT INFORMAT OURCE//HLTH SCI TECHNOL ICIR |
| 2 | 0.0000152608 | MKIAA//SMART CDNA LIBRARY//CDNA SEQUENCING |
| 3 | 0.0000148954 | LATENT PERIODICITY//3 BASE PERIODICITY//DNA WALK |
| 4 | 0.0000116794 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 5 | 0.0000115272 | PROCESSED PSEUDOGENES//PHYSICAL AND PSYCHOLOGICAL STRESSES//DUPLICATED PSEUDOGENES |
| 6 | 0.0000104762 | MULTIPLE SEQUENCE ALIGNMENT//T COFFEE//STATISTICAL ALIGNMENT |
| 7 | 0.0000091487 | MOTIF DISCOVERY//CLOSEST STRING PROBLEM//MOTIF FINDING |
| 8 | 0.0000090755 | CIRCOS//BIODADOS//ORTHOLOGY |
| 9 | 0.0000089520 | STANDARDS IN GENOMIC SCIENCES//BIOL DATA MANAGEMENT TECHNOL//GEBA |
| 10 | 0.0000081131 | WRAP53//OVERLAPPING GENES//CUSTOM BIOTECHNOL SERV GRP |