Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
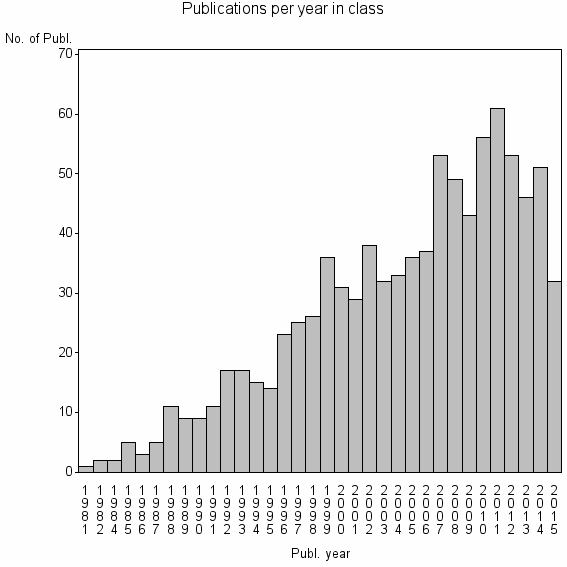
| 11489 | 911 | 49.4 | 91% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 695 | 12311 | CHROMOSOMA//SYNAPTONEMAL COMPLEX//MEIOSIS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CTCF | Author keyword | 50 | 48% | 9% | 78 |
| 2 | CONTROL GENET PROC | Address | 39 | 67% | 4% | 35 |
| 3 | ENHANCER BLOCKING | Author keyword | 31 | 92% | 1% | 12 |
| 4 | SUHW | Author keyword | 29 | 88% | 2% | 14 |
| 5 | BORIS | Author keyword | 21 | 73% | 2% | 16 |
| 6 | FAB 7 | Author keyword | 18 | 89% | 1% | 8 |
| 7 | FAB 8 | Author keyword | 17 | 100% | 1% | 8 |
| 8 | CP190 | Author keyword | 15 | 82% | 1% | 9 |
| 9 | CHROMATIN DOMAIN BOUNDARY | Author keyword | 14 | 100% | 1% | 7 |
| 10 | CHROMATIN INSULATORS | Author keyword | 12 | 86% | 1% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CTCF | 50 | 48% | 9% | 78 | Search CTCF | Search CTCF |
| 2 | ENHANCER BLOCKING | 31 | 92% | 1% | 12 | Search ENHANCER+BLOCKING | Search ENHANCER+BLOCKING |
| 3 | SUHW | 29 | 88% | 2% | 14 | Search SUHW | Search SUHW |
| 4 | BORIS | 21 | 73% | 2% | 16 | Search BORIS | Search BORIS |
| 5 | FAB 7 | 18 | 89% | 1% | 8 | Search FAB+7 | Search FAB+7 |
| 6 | FAB 8 | 17 | 100% | 1% | 8 | Search FAB+8 | Search FAB+8 |
| 7 | CP190 | 15 | 82% | 1% | 9 | Search CP190 | Search CP190 |
| 8 | CHROMATIN DOMAIN BOUNDARY | 14 | 100% | 1% | 7 | Search CHROMATIN+DOMAIN+BOUNDARY | Search CHROMATIN+DOMAIN+BOUNDARY |
| 9 | CHROMATIN INSULATORS | 12 | 86% | 1% | 6 | Search CHROMATIN+INSULATORS | Search CHROMATIN+INSULATORS |
| 10 | GAGA FACTOR | 10 | 52% | 1% | 13 | Search GAGA+FACTOR | Search GAGA+FACTOR |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CHROMATIN DOMAIN BOUNDARY | 137 | 92% | 6% | 55 |
| 2 | HAIRY WING PROTEIN | 118 | 90% | 6% | 52 |
| 3 | ENHANCER BLOCKING ACTIVITY | 83 | 45% | 15% | 138 |
| 4 | ENHANCER BLOCKING | 78 | 61% | 9% | 83 |
| 5 | CHROMATIN INSULATORS | 71 | 72% | 6% | 55 |
| 6 | CHROMATIN INSULATOR | 44 | 47% | 8% | 70 |
| 7 | FAB 7 BOUNDARY | 44 | 100% | 2% | 16 |
| 8 | POLYCOMB RESPONSE ELEMENT | 42 | 59% | 5% | 47 |
| 9 | SUHW INSULATOR | 42 | 94% | 2% | 15 |
| 10 | CIS REGULATORY DOMAINS | 41 | 81% | 3% | 25 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| CTCF: an architectural protein bridging genome topology and function | 2014 | 56 | 99 | 35% |
| CTCF: Master Weaver of the Genome | 2009 | 538 | 99 | 37% |
| We gather together: insulators and genome organization | 2007 | 251 | 47 | 55% |
| Insulators: exploiting transcriptional and epigenetic mechanisms | 2006 | 334 | 83 | 45% |
| Chromatin Insulators: Linking Genome Organization to Cellular Function | 2013 | 35 | 91 | 42% |
| Chromatin Insulators: Regulatory Mechanisms and Epigenetic Inheritance | 2008 | 95 | 64 | 64% |
| Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks | 2014 | 8 | 98 | 73% |
| Does CTCF mediate between nuclear organization and gene expression? | 2010 | 75 | 123 | 42% |
| Gene regulation - Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome | 2001 | 256 | 50 | 60% |
| Chromatin insulators and long-distance interactions in Drosophila | 2014 | 4 | 101 | 90% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CONTROL GENET PROC | 39 | 67% | 3.8% | 35 |
| 2 | GRP TRANSCRIPT REGULAT | 9 | 83% | 0.5% | 5 |
| 3 | MOL PATHOL SECT | 4 | 19% | 2.1% | 19 |
| 4 | GENE EXP S REGULAT DEV | 3 | 100% | 0.3% | 3 |
| 5 | WELLCOME CRC DEV CANC BIOL | 2 | 67% | 0.2% | 2 |
| 6 | DROSOPHILA MOL GENET | 1 | 100% | 0.2% | 2 |
| 7 | EPIGENET REGULAT TRANSCRIPT | 1 | 100% | 0.2% | 2 |
| 8 | LIA 1066 | 1 | 50% | 0.2% | 2 |
| 9 | SECT DROSOPHILA GENE REGULAT | 1 | 100% | 0.2% | 2 |
| 10 | HELEN ROLLASON | 1 | 40% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000164841 | DREF//INSECT BIOMED//DNA REPLICATION RELATED GENE |
| 2 | 0.0000132229 | LOCUS CONTROL REGION//HEMOGLOBIN SWITCHING//EKLF |
| 3 | 0.0000129553 | EZH2//POLYCOMB//POLYCOMB GROUP |
| 4 | 0.0000128819 | CHROMOSOME TERRITORY//CHROMOSOME TERRITORIES//NUCLEAR ARCHITECTURE |
| 5 | 0.0000123864 | P ELEMENT//HYBRID DYSGENESIS//HOBO ELEMENT |
| 6 | 0.0000113115 | HP1//POSITION EFFECT VARIEGATION//HETEROCHROMATIN |
| 7 | 0.0000097499 | CHIP SEQ//PEAK CALLING//DNASE SEQ |
| 8 | 0.0000092639 | BLOOMINGTON DROSOPHILA STOCK//ENDS OUT TARGETING//ENDS OUT |
| 9 | 0.0000073568 | DNA PUFF//DNA PUFFS//RHYNCHOSCIARA |
| 10 | 0.0000064916 | GENOMIC IMPRINTING//H19//BECKWITH WIEDEMANN SYNDROME |