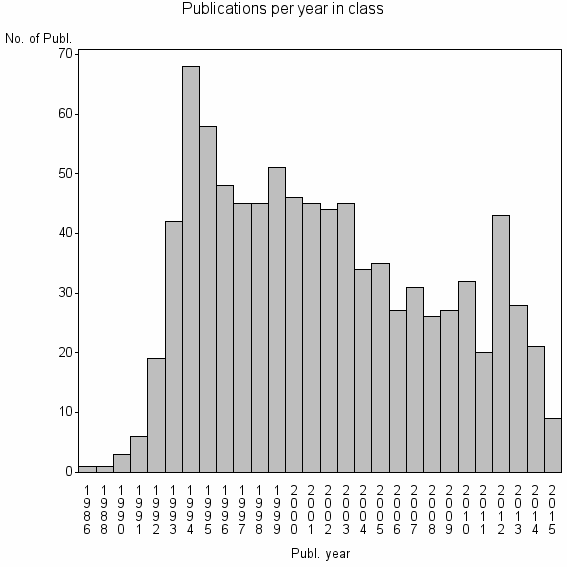
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 11658 | 900 | 44.6 | 89% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 313 | 17635 | X LINKED AGAMMAGLOBULINEMIA//BRUTONS TYROSINE KINASE//BTK |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | GRB2 | Author keyword | 15 | 23% | 6% | 57 |
| 2 | SH2 DOMAIN | Author keyword | 11 | 19% | 6% | 53 |
| 3 | SH3 DOMAIN | Author keyword | 10 | 16% | 7% | 61 |
| 4 | GRB2 SH2 | Author keyword | 9 | 83% | 1% | 5 |
| 5 | GRB2 SH2 DOMAIN | Author keyword | 9 | 83% | 1% | 5 |
| 6 | SH2 | Author keyword | 6 | 20% | 3% | 28 |
| 7 | SH3 DOMAINS | Author keyword | 6 | 29% | 2% | 18 |
| 8 | SH3 | Author keyword | 6 | 17% | 3% | 30 |
| 9 | SH2 DOMAINS | Author keyword | 5 | 27% | 2% | 16 |
| 10 | PHOSPHOTYROSYL MIMETIC | Author keyword | 4 | 75% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GRB2 | 15 | 23% | 6% | 57 | Search GRB2 | Search GRB2 |
| 2 | SH2 DOMAIN | 11 | 19% | 6% | 53 | Search SH2+DOMAIN | Search SH2+DOMAIN |
| 3 | SH3 DOMAIN | 10 | 16% | 7% | 61 | Search SH3+DOMAIN | Search SH3+DOMAIN |
| 4 | GRB2 SH2 | 9 | 83% | 1% | 5 | Search GRB2+SH2 | Search GRB2+SH2 |
| 5 | GRB2 SH2 DOMAIN | 9 | 83% | 1% | 5 | Search GRB2+SH2+DOMAIN | Search GRB2+SH2+DOMAIN |
| 6 | SH2 | 6 | 20% | 3% | 28 | Search SH2 | Search SH2 |
| 7 | SH3 DOMAINS | 6 | 29% | 2% | 18 | Search SH3+DOMAINS | Search SH3+DOMAINS |
| 8 | SH3 | 6 | 17% | 3% | 30 | Search SH3 | Search SH3 |
| 9 | SH2 DOMAINS | 5 | 27% | 2% | 16 | Search SH2+DOMAINS | Search SH2+DOMAINS |
| 10 | PHOSPHOTYROSYL MIMETIC | 4 | 75% | 0% | 3 | Search PHOSPHOTYROSYL+MIMETIC | Search PHOSPHOTYROSYL+MIMETIC |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AFFINITY PHOSPHOTYROSYL PEPTIDE | 70 | 53% | 10% | 92 |
| 2 | SRC HOMOLOGY 2 DOMAIN | 37 | 42% | 8% | 68 |
| 3 | ABL SH3 DOMAIN | 33 | 77% | 3% | 23 |
| 4 | PHOSPHATE CONTAINING LIGANDS | 32 | 78% | 2% | 21 |
| 5 | SH3 LIGAND INTERACTIONS | 27 | 70% | 3% | 23 |
| 6 | NONPHOSPHORYLATED INHIBITOR | 21 | 90% | 1% | 9 |
| 7 | PROLINE RICH PEPTIDES | 20 | 34% | 5% | 48 |
| 8 | TYROSINE PHOSPHORYLATED PEPTIDES | 15 | 48% | 3% | 23 |
| 9 | GRB2 SH2 DOMAIN | 13 | 46% | 2% | 22 |
| 10 | PHOSPHOTYROSINE RECOGNITION | 13 | 71% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Modular peptide recognition domains in eukaryotic signaling | 1997 | 388 | 99 | 51% |
| Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction | 2005 | 221 | 101 | 30% |
| PROTEIN MODULES AND SIGNALING NETWORKS | 1995 | 2017 | 111 | 24% |
| The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains | 2000 | 775 | 137 | 22% |
| MODULAR BINDING DOMAINS IN SIGNAL-TRANSDUCTION PROTEINS | 1995 | 875 | 66 | 41% |
| The SH2 domain: versatile signaling module and pharmaceutical target | 2005 | 126 | 254 | 35% |
| SH3 domain ligand binding: What's the consensus and where's the specificity? | 2012 | 23 | 28 | 46% |
| Can we infer peptide recognition specificity mediated by SH3 domains? | 2002 | 90 | 28 | 64% |
| Evolving specificity from variability for protein interaction domains | 2011 | 15 | 71 | 41% |
| Phosphotyrosyl mimetics in the development of signal transduction inhibitors | 2003 | 70 | 74 | 43% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | VAKGRP COMP WETEN PEN | 2 | 67% | 0.2% | 2 |
| 2 | RAYMOND BEVERLY SACKLER GENET MOL MED | 2 | 26% | 0.8% | 7 |
| 3 | UM SYLVESTER BRAMAN FAMILY BREAST CANC | 2 | 43% | 0.3% | 3 |
| 4 | TEXAS DRUG DIAGNOST DEV | 2 | 21% | 0.8% | 7 |
| 5 | GWEN JULES KN P BIOMED DISCOVERY | 1 | 100% | 0.2% | 2 |
| 6 | UMR CNRS 8600 | 1 | 50% | 0.2% | 2 |
| 7 | VET ADM FDN | 1 | 100% | 0.2% | 2 |
| 8 | FRE 2718 | 1 | 33% | 0.3% | 3 |
| 9 | PHYS CHEM BIOCHEM INORGAN CHEM | 1 | 25% | 0.4% | 4 |
| 10 | STRUCT BIOL BIOMED SCI | 1 | 30% | 0.3% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000205727 | P66SHC//SHC//SHC FAMILY |
| 2 | 0.0000171510 | BIOL CHEMBUNKYO KU//DEMETHOXYVIRIDIN//2 4 MORPHOLINYL 8 PHENYLCHROMONE |
| 3 | 0.0000163021 | PHOSPHOPEPTIDE SYNTHESIS//DI T BUTYL N N DIETHYLPHOSPHORAMIDITE//GLOBAL PHOSPHORYLATION |
| 4 | 0.0000143182 | GRB7//GRB14//GRB10 |
| 5 | 0.0000140303 | NEDD9//HEF1//CRK |
| 6 | 0.0000107326 | SLP 76//LINKER FOR ACTIVATION OF T CELLS//SECT LYMPHOCYTE SIGNALING |
| 7 | 0.0000089456 | SHP 2//SHP 1//GAB1 |
| 8 | 0.0000085405 | SYK//SPLEEN TYROSINE KINASE//R788 |
| 9 | 0.0000084486 | EPS8//MTSS1//I BAR |
| 10 | 0.0000083480 | VAV//VAV1//VAV3 |