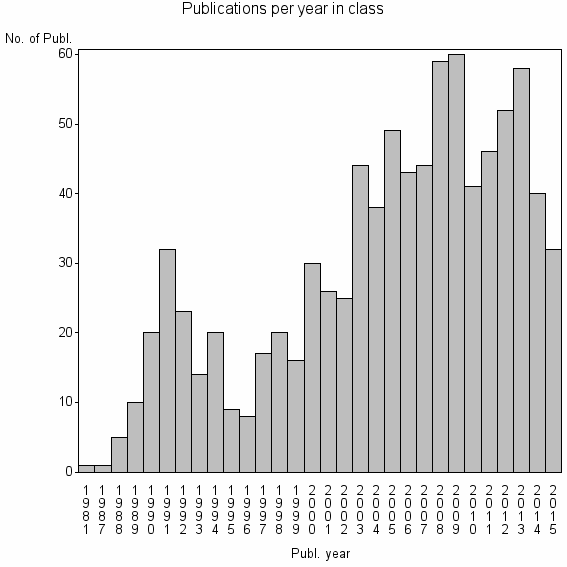
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 11874 | 883 | 48.3 | 94% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1710 | 6112 | FARNESYLTRANSFERASE//PALMITOYLATION//RLIP76 |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RAP1 | Author keyword | 44 | 37% | 11% | 95 |
| 2 | EPAC | Author keyword | 24 | 36% | 6% | 54 |
| 3 | EPAC1 | Author keyword | 21 | 69% | 2% | 18 |
| 4 | RAP2 | Author keyword | 11 | 67% | 1% | 10 |
| 5 | EXCHANGE PROTEIN DIRECTLY ACTIVATED BY CAMP EPAC | Author keyword | 10 | 63% | 1% | 10 |
| 6 | PDZ GEF | Author keyword | 6 | 80% | 0% | 4 |
| 7 | RAP GTPASE | Author keyword | 6 | 80% | 0% | 4 |
| 8 | MOL CANC CANC GENOM NETHERLANDS | Address | 4 | 67% | 0% | 4 |
| 9 | 8 PCPT 2 O ME CAMP | Author keyword | 4 | 75% | 0% | 3 |
| 10 | RAP1 GTPASE | Author keyword | 4 | 75% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RAP1 | 44 | 37% | 11% | 95 | Search RAP1 | Search RAP1 |
| 2 | EPAC | 24 | 36% | 6% | 54 | Search EPAC | Search EPAC |
| 3 | EPAC1 | 21 | 69% | 2% | 18 | Search EPAC1 | Search EPAC1 |
| 4 | RAP2 | 11 | 67% | 1% | 10 | Search RAP2 | Search RAP2 |
| 5 | EXCHANGE PROTEIN DIRECTLY ACTIVATED BY CAMP EPAC | 10 | 63% | 1% | 10 | Search EXCHANGE+PROTEIN+DIRECTLY+ACTIVATED+BY+CAMP+EPAC | Search EXCHANGE+PROTEIN+DIRECTLY+ACTIVATED+BY+CAMP+EPAC |
| 6 | PDZ GEF | 6 | 80% | 0% | 4 | Search PDZ+GEF | Search PDZ+GEF |
| 7 | RAP GTPASE | 6 | 80% | 0% | 4 | Search RAP+GTPASE | Search RAP+GTPASE |
| 8 | 8 PCPT 2 O ME CAMP | 4 | 75% | 0% | 3 | Search 8+PCPT+2++O+ME+CAMP | Search 8+PCPT+2++O+ME+CAMP |
| 9 | RAP1 GTPASE | 4 | 75% | 0% | 3 | Search RAP1+GTPASE | Search RAP1+GTPASE |
| 10 | RAP1A | 2 | 29% | 1% | 6 | Search RAP1A | Search RAP1A |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RAP1 | 41 | 26% | 15% | 136 |
| 2 | SMG P21 | 36 | 77% | 3% | 24 |
| 3 | PUTATIVE EFFECTOR DOMAIN | 35 | 67% | 4% | 32 |
| 4 | EPAC | 21 | 34% | 6% | 50 |
| 5 | SPA 1 DEFICIENT MICE | 19 | 80% | 1% | 12 |
| 6 | EXCHANGE FACTOR EPAC2 | 18 | 83% | 1% | 10 |
| 7 | SMALL GTPASE RAP1 | 15 | 50% | 2% | 21 |
| 8 | MEDIATES RAP1 INDUCED ADHESION | 14 | 41% | 3% | 27 |
| 9 | CAMP EPAC | 12 | 86% | 1% | 6 |
| 10 | TRANSFORMATION SUPPRESSOR ACTIVITY | 11 | 100% | 1% | 6 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Epac: Defining a New Mechanism for cAMP Action | 2010 | 195 | 139 | 57% |
| Epac proteins: multi-purpose cAMP targets | 2006 | 279 | 53 | 72% |
| Linking Rap to cell adhesion | 2005 | 286 | 39 | 67% |
| Rap-linked cAMP signaling Epac proteins: Compartmentation, functioning and disease implications | 2011 | 45 | 157 | 68% |
| Epac: a new cAMP target and new avenues in cAMP research | 2003 | 288 | 38 | 76% |
| Exchange Protein Directly Activated by cAMP (epac): A Multidomain cAMP Mediator in the Regulation of Diverse Biological Functions | 2013 | 31 | 410 | 36% |
| Cyclic AMP sensor EPAC proteins and energy homeostasis | 2014 | 6 | 103 | 39% |
| Regulating Rap small G-proteins in time and space | 2011 | 40 | 89 | 51% |
| Rap1 signalling: Adhering to new models | 2001 | 386 | 94 | 56% |
| The role of Epac proteins, novel cAMP mediators, in the regulation of immune, lung and neuronal function | 2010 | 60 | 210 | 46% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL CANC CANC GENOM NETHERLANDS | 4 | 67% | 0.5% | 4 |
| 2 | SUSAN LEHMAN CULLEN CANC | 4 | 75% | 0.3% | 3 |
| 3 | UCCC DNA SEQUENCING ANAL CORE | 3 | 100% | 0.3% | 3 |
| 4 | BAYER CHAIR MOL IMMUNOL ALLERGY | 3 | 60% | 0.3% | 3 |
| 5 | MOL IMMUNOL ALLERGY | 2 | 44% | 0.5% | 4 |
| 6 | CELLULAR MOL BIOL REPROD | 2 | 67% | 0.2% | 2 |
| 7 | UMR S769 | 2 | 31% | 0.6% | 5 |
| 8 | BIOMED GENET CANC GENOM | 1 | 100% | 0.2% | 2 |
| 9 | DEV NEUROBIOLCHUO KU | 1 | 50% | 0.2% | 2 |
| 10 | I3 GRP | 1 | 50% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000150513 | RLIP76//RALBP1//RAL |
| 2 | 0.0000127285 | NEDD9//HEF1//CRK |
| 3 | 0.0000101502 | GRAVIN//AKAP//A KINASE ANCHORING PROTEIN |
| 4 | 0.0000092014 | FARNESYLTHIOSALICYLIC ACID//SALIRASIB//ABT STRUKTURELLE BIOL |
| 5 | 0.0000076192 | STE20 LIKE KINASE//MAP4K4//STE20 |
| 6 | 0.0000072417 | ONCOGENIC RAS P21//EFFECTOR DOMAINS//N RAS |
| 7 | 0.0000071243 | RAF//RAF 1//LEO JENKINS CANC |
| 8 | 0.0000062533 | DIACYLGLYCEROL KINASE//DIACYLGLYCEROL KINASE DGK//DGK ZETA |
| 9 | 0.0000060492 | SECT PULM CRIT MED//LUNG INJURY//ENDOTHELIAL PERMEABILITY |
| 10 | 0.0000055282 | CAPK//CAMP DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT//PATHOCHEM |