Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
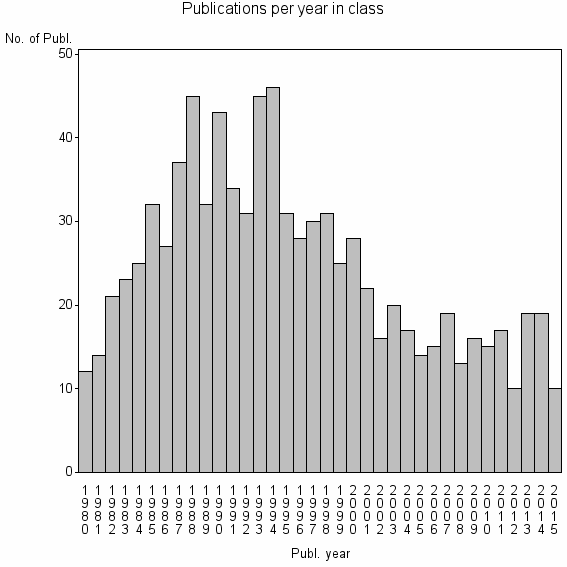
| 11886 | 882 | 40.5 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 134 | 22710 | SACCHAROMYCES CEREVISIAE//YEAST//SCHIZOSACCHAROMYCES POMBE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CDC25MM | Author keyword | 12 | 86% | 1% | 6 |
| 2 | RAS GRF1 | Author keyword | 10 | 73% | 1% | 8 |
| 3 | BCY1 | Author keyword | 7 | 64% | 1% | 7 |
| 4 | RAS GRF | Author keyword | 7 | 64% | 1% | 7 |
| 5 | ADENYLYL CYCLASE ASSOCIATED PROTEIN | Author keyword | 6 | 80% | 0% | 4 |
| 6 | SRV2 | Author keyword | 6 | 80% | 0% | 4 |
| 7 | CYCLASE ASSOCIATED PROTEIN | Author keyword | 6 | 58% | 1% | 7 |
| 8 | INFORMAT GENET DEV | Address | 6 | 58% | 1% | 7 |
| 9 | MSN2P | Author keyword | 4 | 50% | 1% | 6 |
| 10 | RASGRF | Author keyword | 4 | 75% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CDC25MM | 12 | 86% | 1% | 6 | Search CDC25MM | Search CDC25MM |
| 2 | RAS GRF1 | 10 | 73% | 1% | 8 | Search RAS+GRF1 | Search RAS+GRF1 |
| 3 | BCY1 | 7 | 64% | 1% | 7 | Search BCY1 | Search BCY1 |
| 4 | RAS GRF | 7 | 64% | 1% | 7 | Search RAS+GRF | Search RAS+GRF |
| 5 | ADENYLYL CYCLASE ASSOCIATED PROTEIN | 6 | 80% | 0% | 4 | Search ADENYLYL+CYCLASE+ASSOCIATED+PROTEIN | Search ADENYLYL+CYCLASE+ASSOCIATED+PROTEIN |
| 6 | SRV2 | 6 | 80% | 0% | 4 | Search SRV2 | Search SRV2 |
| 7 | CYCLASE ASSOCIATED PROTEIN | 6 | 58% | 1% | 7 | Search CYCLASE+ASSOCIATED+PROTEIN | Search CYCLASE+ASSOCIATED+PROTEIN |
| 8 | MSN2P | 4 | 50% | 1% | 6 | Search MSN2P | Search MSN2P |
| 9 | RASGRF | 4 | 75% | 0% | 3 | Search RASGRF | Search RASGRF |
| 10 | CAP1 | 4 | 41% | 1% | 7 | Search CAP1 | Search CAP1 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CDC25 GENE PRODUCT | 32 | 80% | 2% | 20 |
| 2 | CDC25 GENE | 22 | 58% | 3% | 26 |
| 3 | SDC25 C DOMAIN | 18 | 89% | 1% | 8 |
| 4 | SACCHAROMYCES CEREVISIAE CDC25 | 15 | 68% | 1% | 13 |
| 5 | PROTEIN MEDIATED SIGNALS | 12 | 75% | 1% | 9 |
| 6 | ADENYLATE CYCLASE PATHWAY | 12 | 59% | 1% | 13 |
| 7 | CYCLASE PATHWAY | 9 | 50% | 1% | 13 |
| 8 | CYCLASE ASSOCIATED PROTEIN | 9 | 26% | 3% | 29 |
| 9 | IRA2 | 8 | 75% | 1% | 6 |
| 10 | KELCH REPEAT PROTEINS | 8 | 45% | 1% | 13 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae | 1999 | 379 | 94 | 50% |
| Molecular mechanisms of feedback inhibition of protein kinase A on intracellular cAMP accumulation | 2012 | 20 | 152 | 49% |
| The RasGrf family of mammalian guanine nucleotide exchange factors | 2011 | 25 | 155 | 43% |
| Many faces of Ras activation | 2008 | 74 | 150 | 33% |
| STATIONARY-PHASE IN THE YEAST SACCHAROMYCES-CEREVISIAE | 1993 | 406 | 203 | 31% |
| Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae | 2007 | 37 | 73 | 44% |
| Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae | 2010 | 79 | 386 | 18% |
| Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization | 2002 | 106 | 74 | 45% |
| SIGNAL-TRANSDUCTION IN YEAST | 1994 | 251 | 209 | 22% |
| The role of mitochondrial biogenesis and ROS in the control of energy supply in proliferating cells | 2014 | 2 | 54 | 24% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | INFORMAT GENET DEV | 6 | 58% | 0.8% | 7 |
| 2 | GRP BIOPHYS EQUIPE 2 | 2 | 50% | 0.3% | 3 |
| 3 | MOL CELBIOL | 2 | 11% | 1.7% | 15 |
| 4 | COTTON GERMPLASM | 1 | 100% | 0.2% | 2 |
| 5 | IBMCC CSIC USAL | 1 | 33% | 0.3% | 3 |
| 6 | MARKEY CELL SIGNALLING | 1 | 40% | 0.2% | 2 |
| 7 | SEZ BIOCHIM COMPARATA | 1 | 18% | 0.5% | 4 |
| 8 | 11 MINAMIOOYA | 1 | 50% | 0.1% | 1 |
| 9 | BIOCHEM POB 20151 | 1 | 50% | 0.1% | 1 |
| 10 | BIOTECHNOL NEGEV NIBN | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000154160 | IME1//IME2//PROSPORE MEMBRANE |
| 2 | 0.0000131921 | GLUCOSE REPRESSION//CATABOLITE INACTIVATION//MIG1 |
| 3 | 0.0000126530 | TREHALASE//TREHALOSE//TREHALOSE 6 PHOSPHATE SYNTHASE |
| 4 | 0.0000123031 | ONCOGENIC RAS P21//EFFECTOR DOMAINS//N RAS |
| 5 | 0.0000110696 | SPORANGIOSPORES//YEASTLIKE CELLS//BENJAMINIELLA POITRASII |
| 6 | 0.0000108148 | FARNESYLTHIOSALICYLIC ACID//SALIRASIB//ABT STRUKTURELLE BIOL |
| 7 | 0.0000095800 | FLO11//FLO1//FLO GENES |
| 8 | 0.0000093153 | CHRONOLOGICAL LIFESPAN//YEAST APOPTOSIS//RETROGRADE RESPONSE |
| 9 | 0.0000084526 | RSP5 UBIQUITIN LIGASE//GLN3//RSP5 |
| 10 | 0.0000078701 | PHEROMONE RESPONSE//STE2//CDC42P |