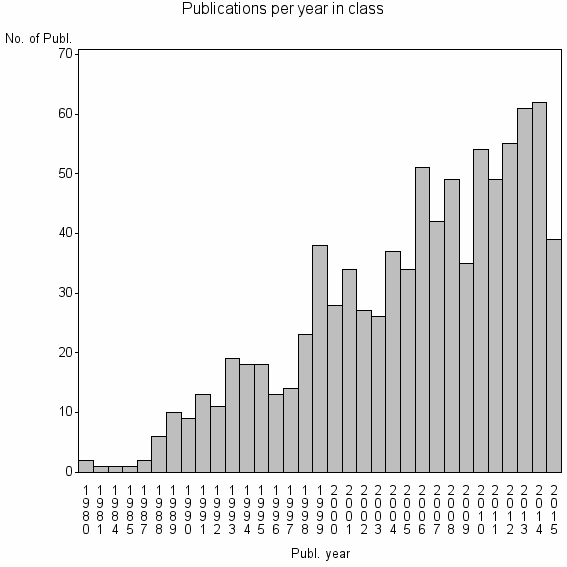
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 11891 | 882 | 46.8 | 91% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RNA HELICASE | Author keyword | 46 | 37% | 12% | 102 |
| 2 | DEAD BOX | Author keyword | 46 | 64% | 5% | 45 |
| 3 | DEAD BOX PROTEIN | Author keyword | 34 | 58% | 4% | 39 |
| 4 | DEAD BOX RNA HELICASE | Author keyword | 25 | 60% | 3% | 27 |
| 5 | RNA UNWINDING | Author keyword | 24 | 82% | 2% | 14 |
| 6 | DDX3 | Author keyword | 23 | 64% | 3% | 23 |
| 7 | P68 RNA HELICASE | Author keyword | 23 | 86% | 1% | 12 |
| 8 | UNWINDING ENZYME | Author keyword | 15 | 71% | 1% | 12 |
| 9 | P68 | Author keyword | 14 | 57% | 2% | 17 |
| 10 | PLANT DNA HELICASE | Author keyword | 12 | 86% | 1% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA HELICASE | 46 | 37% | 12% | 102 | Search RNA+HELICASE | Search RNA+HELICASE |
| 2 | DEAD BOX | 46 | 64% | 5% | 45 | Search DEAD+BOX | Search DEAD+BOX |
| 3 | DEAD BOX PROTEIN | 34 | 58% | 4% | 39 | Search DEAD+BOX+PROTEIN | Search DEAD+BOX+PROTEIN |
| 4 | DEAD BOX RNA HELICASE | 25 | 60% | 3% | 27 | Search DEAD+BOX+RNA+HELICASE | Search DEAD+BOX+RNA+HELICASE |
| 5 | RNA UNWINDING | 24 | 82% | 2% | 14 | Search RNA+UNWINDING | Search RNA+UNWINDING |
| 6 | DDX3 | 23 | 64% | 3% | 23 | Search DDX3 | Search DDX3 |
| 7 | P68 RNA HELICASE | 23 | 86% | 1% | 12 | Search P68+RNA+HELICASE | Search P68+RNA+HELICASE |
| 8 | UNWINDING ENZYME | 15 | 71% | 1% | 12 | Search UNWINDING+ENZYME | Search UNWINDING+ENZYME |
| 9 | P68 | 14 | 57% | 2% | 17 | Search P68 | Search P68 |
| 10 | PLANT DNA HELICASE | 12 | 86% | 1% | 6 | Search PLANT+DNA+HELICASE | Search PLANT+DNA+HELICASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ESCHERICHIA COLI DBPA | 49 | 94% | 2% | 17 |
| 2 | FACTOR EIF 4A | 37 | 63% | 4% | 37 |
| 3 | INITIATION FACTOR 4A | 32 | 38% | 8% | 67 |
| 4 | FACTOR 4A | 27 | 65% | 3% | 26 |
| 5 | DEAD BOX PROTEINS | 23 | 40% | 5% | 46 |
| 6 | DEAD BOX PROTEIN | 21 | 31% | 6% | 56 |
| 7 | PROTEIN DBPA | 20 | 100% | 1% | 9 |
| 8 | A D BOX | 19 | 68% | 2% | 17 |
| 9 | GENERAL RNA | 18 | 83% | 1% | 10 |
| 10 | EUKARYOTIC INITIATION FACTORS | 15 | 47% | 3% | 23 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| From unwinding to clamping - the DEAD box RNA helicase family | 2011 | 164 | 137 | 59% |
| The DEAD-box protein family of RNA helicases | 2006 | 443 | 151 | 55% |
| RNA Helicase Proteins as Chaperones and Remodelers | 2014 | 13 | 232 | 43% |
| SF1 and SF2 helicases: family matters | 2010 | 198 | 89 | 27% |
| The DEAD box proteins DDX5 (p68) and DDX17 (p72): Multi-tasking transcriptional regulators | 2013 | 14 | 63 | 84% |
| mRNA helicases: the tacticians of translational control | 2011 | 97 | 115 | 51% |
| Dead-box proteins: The driving forces behind RNA metabolism | 2004 | 383 | 94 | 46% |
| Looking back on the birth of DEAD-box RNA helicases | 2013 | 15 | 82 | 77% |
| RNA helicases at work: binding and rearranging | 2011 | 117 | 97 | 42% |
| DEAD-box helicases as integrators of RNA, nucleotide and protein binding | 2013 | 16 | 108 | 72% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SECT MOL ENDOCRINOL | 5 | 28% | 1.7% | 15 |
| 2 | HAEMATOPATHOL | 2 | 17% | 1.2% | 11 |
| 3 | MICROBIOL MED MOL | 1 | 10% | 1.0% | 9 |
| 4 | STRUCT GEN CONSORTIUM | 1 | 33% | 0.2% | 2 |
| 5 | BIOCHIM ENZYMOL STRUCT | 1 | 50% | 0.1% | 1 |
| 6 | CNRSUMR7212INSERMU944 | 1 | 50% | 0.1% | 1 |
| 7 | KONERU LAKSHMAIAH EDUC FDN | 1 | 50% | 0.1% | 1 |
| 8 | MED FOOD | 1 | 50% | 0.1% | 1 |
| 9 | MED OURCE UNIT | 1 | 50% | 0.1% | 1 |
| 10 | MEULLER 208 | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000137512 | OBSTET GYNECOLSEALY STRUCT BIOL//PRIMASE//HELICASE |
| 2 | 0.0000131834 | EIF4E//EIF 4E//CAPPING EFFICIENCY |
| 3 | 0.0000108623 | PDCD4//PROGRAMMED CELL DEATH 4//PROGRAMMED CELL DEATH 4 PDCD4 |
| 4 | 0.0000093082 | DECAPPING//P BODIES//CCR4 NOT |
| 5 | 0.0000088241 | UPF1//NMD//NONSENSE MEDIATED MRNA DECAY |
| 6 | 0.0000077124 | SPLICEOSOME//PRE MRNA SPLICING//PRP24 |
| 7 | 0.0000075651 | MRNP BIOGENESIS METAB//DIS3//RRP6 |
| 8 | 0.0000072432 | COLD SHOCK PROTEIN//GLYCINE RICH RNA BINDING PROTEIN//CIRP |
| 9 | 0.0000071377 | REV//RRE//REV PROTEIN |
| 10 | 0.0000068370 | SNORNA//SMALL NUCLEOLAR RNAS//PRE RRNA PROCESSING |