Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
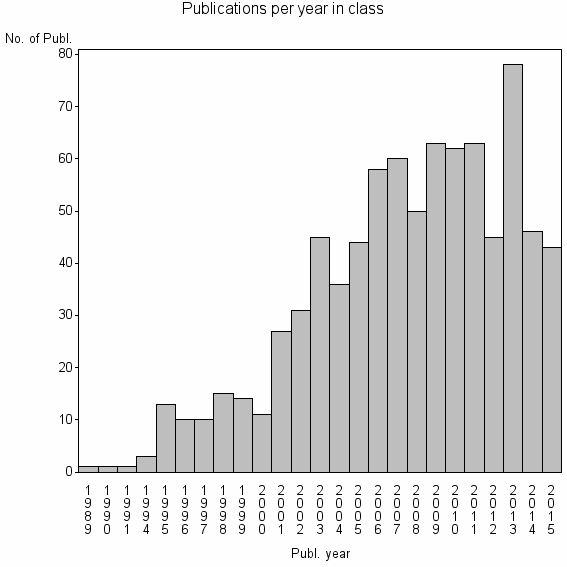
| 12618 | 830 | 46.1 | 85% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1180 | 8915 | ANKYLOSING SPONDYLITIS//HLA B27//SPONDYLOARTHRITIS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | IMMUNOINFORMATICS | Author keyword | 26 | 47% | 5% | 41 |
| 2 | EPITOPE PREDICTION | Author keyword | 20 | 38% | 5% | 41 |
| 3 | COMPUTATIONAL VACCINOLOGY | Author keyword | 10 | 73% | 1% | 8 |
| 4 | SUPERTYPES | Author keyword | 8 | 70% | 1% | 7 |
| 5 | PEPTIDE BINDING PREDICTION | Author keyword | 8 | 100% | 1% | 5 |
| 6 | DEIMMUNIZATION | Author keyword | 8 | 62% | 1% | 8 |
| 7 | TB HIV | Address | 7 | 44% | 1% | 12 |
| 8 | MHC BINDING | Author keyword | 7 | 57% | 1% | 8 |
| 9 | T CELL EPITOPE PREDICTION | Author keyword | 6 | 58% | 1% | 7 |
| 10 | HLA A0201 | Author keyword | 5 | 26% | 2% | 17 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | IMMUNOINFORMATICS | 26 | 47% | 5% | 41 | Search IMMUNOINFORMATICS | Search IMMUNOINFORMATICS |
| 2 | EPITOPE PREDICTION | 20 | 38% | 5% | 41 | Search EPITOPE+PREDICTION | Search EPITOPE+PREDICTION |
| 3 | COMPUTATIONAL VACCINOLOGY | 10 | 73% | 1% | 8 | Search COMPUTATIONAL+VACCINOLOGY | Search COMPUTATIONAL+VACCINOLOGY |
| 4 | SUPERTYPES | 8 | 70% | 1% | 7 | Search SUPERTYPES | Search SUPERTYPES |
| 5 | PEPTIDE BINDING PREDICTION | 8 | 100% | 1% | 5 | Search PEPTIDE+BINDING+PREDICTION | Search PEPTIDE+BINDING+PREDICTION |
| 6 | DEIMMUNIZATION | 8 | 62% | 1% | 8 | Search DEIMMUNIZATION | Search DEIMMUNIZATION |
| 7 | MHC BINDING | 7 | 57% | 1% | 8 | Search MHC+BINDING | Search MHC+BINDING |
| 8 | T CELL EPITOPE PREDICTION | 6 | 58% | 1% | 7 | Search T+CELL+EPITOPE+PREDICTION | Search T+CELL+EPITOPE+PREDICTION |
| 9 | HLA A0201 | 5 | 26% | 2% | 17 | Search HLA+A0201 | Search HLA+A0201 |
| 10 | BINDING PREDICTION | 5 | 63% | 1% | 5 | Search BINDING+PREDICTION | Search BINDING+PREDICTION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROTEASOMAL CLEAVAGE | 37 | 67% | 4% | 34 |
| 2 | TAP TRANSPORT EFFICIENCY | 31 | 73% | 3% | 24 |
| 3 | SUPERTYPES | 28 | 64% | 3% | 28 |
| 4 | T CELL EPITOPES | 26 | 15% | 18% | 153 |
| 5 | NETMHCPAN | 24 | 91% | 1% | 10 |
| 6 | INDEPENDENT BINDING | 23 | 65% | 3% | 22 |
| 7 | MHC BINDING | 21 | 45% | 4% | 35 |
| 8 | PEPTIDE BINDING PREDICTION | 21 | 85% | 1% | 11 |
| 9 | CLASS I BINDING | 19 | 55% | 3% | 24 |
| 10 | MOLECULE HLA A ASTERISK 0201 | 18 | 83% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools | 2012 | 21 | 58 | 81% |
| T-cell epitope vaccine design by immunoinformatics | 2013 | 18 | 138 | 38% |
| Prediction of MHC-Peptide Binding: A Systematic and Comprehensive Overview | 2009 | 40 | 105 | 65% |
| Predicting peptide binding to Major Histocompatibility Complex molecules | 2011 | 15 | 38 | 68% |
| An introduction to epitope prediction methods and software | 2009 | 39 | 116 | 59% |
| HLA class I supertypes: a revised and updated classification | 2008 | 145 | 107 | 36% |
| MHC Class II epitope predictive algorithms | 2010 | 37 | 90 | 66% |
| Reverse Vaccinology: Developing Vaccines in the Era of Genomics | 2010 | 111 | 123 | 24% |
| Major histocompatibility complex class I binding predictions as a tool in epitope discovery | 2010 | 29 | 92 | 61% |
| Prediction of Immunogenicity of Therapeutic Proteins Validity of Computational Tools | 2010 | 31 | 35 | 51% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TB HIV | 7 | 44% | 1.4% | 12 |
| 2 | IMMUNOL INFORMAT | 5 | 24% | 2.0% | 17 |
| 3 | SVIMS BIOINFORMAT | 3 | 60% | 0.4% | 3 |
| 4 | WSI ZBIT | 3 | 60% | 0.4% | 3 |
| 5 | KENT RIDGE DIGITAL S | 2 | 40% | 0.5% | 4 |
| 6 | PROD EVALUAT REGISTRAT | 1 | 50% | 0.2% | 2 |
| 7 | UNIT ANIM MED COMPUTAT MED BIOINFORMAT | 1 | 100% | 0.2% | 2 |
| 8 | SIMULAT BIOL SYST | 1 | 20% | 0.7% | 6 |
| 9 | THEORY NEC | 1 | 40% | 0.2% | 2 |
| 10 | IMMUNOMED | 1 | 33% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000192868 | TAPASIN//TRANSPORTER ASSOCIATED WITH ANTIGEN PROCESSING//PEPTIDE LOADING COMPLEX |
| 2 | 0.0000114358 | PREDETERMINED SPECIFICITY//SYNTHETIC ANTIGENIC SITE//B CELL EPITOPE PREDICTION |
| 3 | 0.0000091748 | INVARIANT CHAIN//HLA DM//ANTIGEN PROCESSING |
| 4 | 0.0000087415 | PARTNERS AIDS//HIV PATHOGENESIS PROGRAMME//DORIS DUKE MED |
| 5 | 0.0000081665 | PEPTIDE MHC//PEPTIDE MAJOR HISTOCOMPATIBILITY COMPLEX//PROT CRYSTALLOG UNIT |
| 6 | 0.0000076400 | CANC VACCINE DEV PROGRAM//CANC SECT//ONCOANTIGENS |
| 7 | 0.0000075930 | PROGRAM TRANSLAT IMMUNOVIROL BIODEF//MAYO VACCINE GRP//MAYO CLIN VACCINE GRP |
| 8 | 0.0000064134 | CANCER TESTIS ANTIGEN//CANCER TESTIS ANTIGENS//NY ESO 1 |
| 9 | 0.0000057947 | CANC VACCINE DEV//INNOVAT CANC THER Y//PERSONALIZED PEPTIDE VACCINE |
| 10 | 0.0000049057 | MINOR HISTOCOMPATIBILITY ANTIGENS//MINOR HISTOCOMPATIBILITY ANTIGEN//HA 1 |