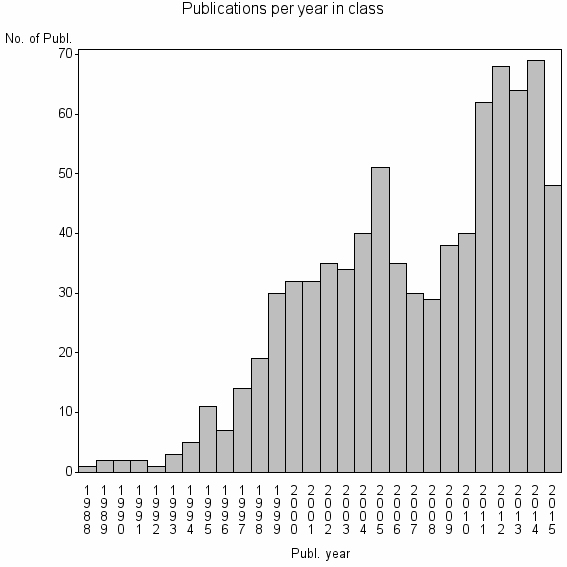
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 12983 | 804 | 49.0 | 94% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MED12 | Author keyword | 82 | 88% | 5% | 38 |
| 2 | MEDIATOR COMPLEX | Author keyword | 54 | 83% | 4% | 30 |
| 3 | MED19 | Author keyword | 27 | 92% | 1% | 11 |
| 4 | MED13 | Author keyword | 14 | 100% | 1% | 7 |
| 5 | CDK8 | Author keyword | 12 | 50% | 2% | 18 |
| 6 | TRAP220 | Author keyword | 11 | 100% | 1% | 6 |
| 7 | CREAT INITIAT GENOME REGULAT | Address | 9 | 83% | 1% | 5 |
| 8 | MED23 | Author keyword | 8 | 100% | 1% | 5 |
| 9 | CYCLIN C | Author keyword | 6 | 44% | 1% | 11 |
| 10 | HOPA | Author keyword | 6 | 100% | 0% | 4 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MED12 | 82 | 88% | 5% | 38 | Search MED12 | Search MED12 |
| 2 | MEDIATOR COMPLEX | 54 | 83% | 4% | 30 | Search MEDIATOR+COMPLEX | Search MEDIATOR+COMPLEX |
| 3 | MED19 | 27 | 92% | 1% | 11 | Search MED19 | Search MED19 |
| 4 | MED13 | 14 | 100% | 1% | 7 | Search MED13 | Search MED13 |
| 5 | CDK8 | 12 | 50% | 2% | 18 | Search CDK8 | Search CDK8 |
| 6 | TRAP220 | 11 | 100% | 1% | 6 | Search TRAP220 | Search TRAP220 |
| 7 | MED23 | 8 | 100% | 1% | 5 | Search MED23 | Search MED23 |
| 8 | CYCLIN C | 6 | 44% | 1% | 11 | Search CYCLIN+C | Search CYCLIN+C |
| 9 | HOPA | 6 | 100% | 0% | 4 | Search HOPA | Search HOPA |
| 10 | KOHTALO | 6 | 100% | 0% | 4 | Search KOHTALO | Search KOHTALO |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | YEAST MEDIATOR | 83 | 90% | 4% | 36 |
| 2 | COFACTOR COMPLEX | 81 | 87% | 5% | 40 |
| 3 | COACTIVATOR COMPLEX | 79 | 54% | 13% | 101 |
| 4 | MEDIATOR COMPLEX | 46 | 48% | 9% | 71 |
| 5 | HUMAN CDK8 SUBCOMPLEX | 44 | 88% | 3% | 21 |
| 6 | TRAP220 COMPONENT | 35 | 67% | 4% | 32 |
| 7 | MAMMALIAN MEDIATOR | 32 | 85% | 2% | 17 |
| 8 | HEAD MODULE | 31 | 82% | 2% | 18 |
| 9 | OPITZ KAVEGGIA SYNDROME | 29 | 88% | 2% | 14 |
| 10 | TRAP220 | 21 | 90% | 1% | 9 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation | 2010 | 221 | 124 | 77% |
| Function and regulation of the Mediator complex | 2011 | 123 | 53 | 91% |
| The Mediator complex: a central integrator of transcription | 2015 | 4 | 194 | 55% |
| The human Mediator complex: a versatile, genome-wide regulator of transcription | 2010 | 137 | 79 | 65% |
| Mediator and the mechanism of transcriptional activation | 2005 | 263 | 31 | 68% |
| The Mediator complex and transcription regulation | 2013 | 41 | 440 | 61% |
| Dynamic regulation of pol II transcription by the mammalian Mediator complex | 2005 | 217 | 78 | 67% |
| The mammalian Mediator complex and its role in transcriptional regulation | 2005 | 157 | 56 | 86% |
| Mechanisms of Mediator complex action in transcriptional activation | 2013 | 23 | 159 | 77% |
| Involvement of Mediator complex in malignancy | 2014 | 11 | 134 | 60% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CREAT INITIAT GENOME REGULAT | 9 | 83% | 0.6% | 5 |
| 2 | HELSINKI UNIV CENT HOSP HUS | 2 | 67% | 0.2% | 2 |
| 3 | DIAGNOST NUCL DEV SDN | 2 | 33% | 0.6% | 5 |
| 4 | IMMUNOCHIM REGULAT CELLULAI INTERACT VIRALES | 1 | 50% | 0.2% | 2 |
| 5 | MOL GENET BRANCH | 1 | 50% | 0.2% | 2 |
| 6 | UOC IMMUNOHEMATOL TRANSFUS MED TRANSPLANT IMMUN | 1 | 27% | 0.4% | 3 |
| 7 | AGIMFRE3405 | 1 | 50% | 0.1% | 1 |
| 8 | BIOCHEM NUCL ACID ENZYMOL | 1 | 50% | 0.1% | 1 |
| 9 | BIOZENTRUM SWISS NANOSCI | 1 | 50% | 0.1% | 1 |
| 10 | CNRS UPS UMR5547 | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000235142 | TFIID//TFIIA//MOT1 |
| 2 | 0.0000128625 | P TEFB//CDK9//HEXIM1 |
| 3 | 0.0000121779 | GAL4P//GAL80//GAL GENES |
| 4 | 0.0000113507 | AIB1//STEROID HORMONES SECT//RIP140 |
| 5 | 0.0000072473 | MYST//MOZ//HISTONE ACETYLTRANSFERASE |
| 6 | 0.0000055918 | SWI SNF//BRG1//ISWI |
| 7 | 0.0000045501 | IME1//IME2//PROSPORE MEMBRANE |
| 8 | 0.0000040592 | HMGA1//HMGA2//HMGIY |
| 9 | 0.0000039922 | WALLEYE DERMAL SARCOMA VIRUS//DERMAL SARCOMA//RETROVIRAL CYCLIN |
| 10 | 0.0000038203 | START//MBF//CDC28 KINASE |