Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
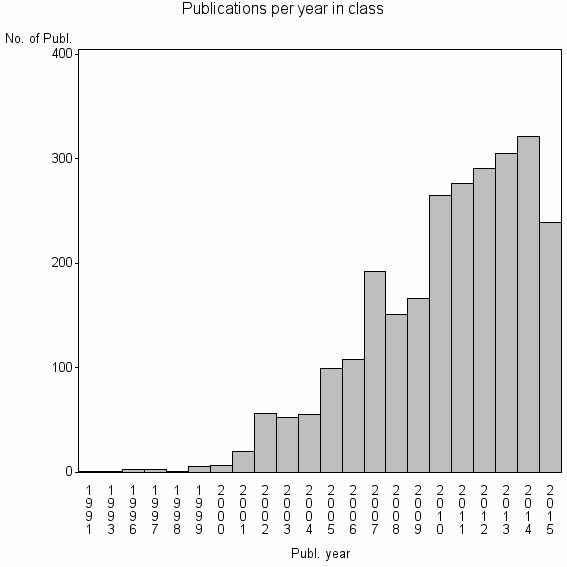
| 1336 | 2614 | 54.3 | 95% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | HISTONE DEMETHYLASE | Author keyword | 100 | 66% | 4% | 93 |
| 2 | HISTONE DEMETHYLATION | Author keyword | 42 | 73% | 1% | 32 |
| 3 | LSD1 | Author keyword | 35 | 52% | 2% | 48 |
| 4 | LYSINE METHYLATION | Author keyword | 30 | 53% | 2% | 40 |
| 5 | HISTONE METHYLATION | Author keyword | 26 | 19% | 5% | 120 |
| 6 | DEMETHYLASE | Author keyword | 22 | 47% | 1% | 34 |
| 7 | JMJC DOMAIN | Author keyword | 21 | 71% | 1% | 17 |
| 8 | PHF8 | Author keyword | 21 | 90% | 0% | 9 |
| 9 | JMJD1A | Author keyword | 20 | 100% | 0% | 9 |
| 10 | LYSINE SPECIFIC DEMETHYLASE | Author keyword | 20 | 100% | 0% | 9 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HISTONE DEMETHYLASE | 100 | 66% | 4% | 93 | Search HISTONE+DEMETHYLASE | Search HISTONE+DEMETHYLASE |
| 2 | HISTONE DEMETHYLATION | 42 | 73% | 1% | 32 | Search HISTONE+DEMETHYLATION | Search HISTONE+DEMETHYLATION |
| 3 | LSD1 | 35 | 52% | 2% | 48 | Search LSD1 | Search LSD1 |
| 4 | LYSINE METHYLATION | 30 | 53% | 2% | 40 | Search LYSINE+METHYLATION | Search LYSINE+METHYLATION |
| 5 | HISTONE METHYLATION | 26 | 19% | 5% | 120 | Search HISTONE+METHYLATION | Search HISTONE+METHYLATION |
| 6 | DEMETHYLASE | 22 | 47% | 1% | 34 | Search DEMETHYLASE | Search DEMETHYLASE |
| 7 | JMJC DOMAIN | 21 | 71% | 1% | 17 | Search JMJC+DOMAIN | Search JMJC+DOMAIN |
| 8 | PHF8 | 21 | 90% | 0% | 9 | Search PHF8 | Search PHF8 |
| 9 | JMJD1A | 20 | 100% | 0% | 9 | Search JMJD1A | Search JMJD1A |
| 10 | LYSINE SPECIFIC DEMETHYLASE | 20 | 100% | 0% | 9 | Search LYSINE+SPECIFIC+DEMETHYLASE | Search LYSINE+SPECIFIC+DEMETHYLASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | LSD1 | 129 | 64% | 5% | 127 |
| 2 | LYSINE DEMETHYLASES | 118 | 95% | 1% | 39 |
| 3 | LYSINE METHYLATION | 90 | 41% | 7% | 170 |
| 4 | H3 | 84 | 27% | 10% | 270 |
| 5 | TRIMETHYLATION | 73 | 46% | 5% | 118 |
| 6 | JMJD2 FAMILY | 70 | 79% | 2% | 45 |
| 7 | CLEFT LIP CLEFT PALATE | 56 | 81% | 1% | 34 |
| 8 | H3 METHYLATION | 49 | 43% | 3% | 86 |
| 9 | LYSINE 4 METHYLATION | 49 | 56% | 2% | 59 |
| 10 | HISTONE DEMETHYLASE | 44 | 64% | 2% | 43 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Chromatin modifications and their function | 2007 | 3808 | 89 | 38% |
| Regulation of chromatin by histone modifications | 2011 | 589 | 121 | 33% |
| Non-histone protein methylation as a regulator of cellular signalling and function | 2015 | 8 | 113 | 36% |
| Molecular mechanisms and potential functions of histone demethylases | 2012 | 158 | 198 | 73% |
| Histone methylation: a dynamic mark in health, disease and inheritance | 2012 | 279 | 179 | 36% |
| The complex language of chromatin regulation during transcription | 2007 | 1163 | 48 | 56% |
| Critical roles of non-histone protein lysine methylation in human tumorigenesis | 2015 | 3 | 154 | 54% |
| Regulation of nucleosome dynamics by histone modifications | 2013 | 159 | 101 | 27% |
| KDM4/JMJD2 Histone Demethylases: Epigenetic Regulators in Cancer Cells | 2013 | 45 | 72 | 81% |
| Histone lysine demethylases as targets for anticancer therapy | 2013 | 48 | 180 | 68% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | STRUCT GENOM CONSORTIUM | 11 | 12% | 3.3% | 85 |
| 2 | ULTZ 415 | 8 | 100% | 0.2% | 5 |
| 3 | OXFORD BIOMED UNIT | 7 | 46% | 0.5% | 12 |
| 4 | CHROMATIN BIOL | 7 | 21% | 1.2% | 31 |
| 5 | INTEGRAT CHEM BIOL DRUG DISCOVERY | 7 | 29% | 0.8% | 20 |
| 6 | TOP DOWN PROTEOM | 6 | 80% | 0.2% | 4 |
| 7 | HISTONE MODIFICAT GRP | 6 | 47% | 0.3% | 9 |
| 8 | INTEGRATED CHEM BIOL DRUG DISCOVERY | 4 | 75% | 0.1% | 3 |
| 9 | NUCLEIC ACIDS ENZYMOL BIOCHEM | 4 | 75% | 0.1% | 3 |
| 10 | CHROMATIN BIOCHEM | 4 | 29% | 0.5% | 12 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000165979 | ARGININE METHYLATION//CARM1//PROTEIN ARGININE METHYLTRANSFERASE |
| 2 | 0.0000127952 | EZH2//POLYCOMB//POLYCOMB GROUP |
| 3 | 0.0000117551 | HISTONE CHAPERONE//ASF1//CAF 1 |
| 4 | 0.0000114601 | HP1//POSITION EFFECT VARIEGATION//HETEROCHROMATIN |
| 5 | 0.0000111271 | METABOLIC MEMORY//HYPERGLYCEMIC MEMORY//EPIGEN PROFILING IL |
| 6 | 0.0000109633 | UHRF1//ICBP90//UHRF2 |
| 7 | 0.0000108751 | MYST//MOZ//HISTONE ACETYLTRANSFERASE |
| 8 | 0.0000092086 | NUT MIDLINE CARCINOMA//BROMODOMAIN//BRD4 |
| 9 | 0.0000085644 | CHIP SEQ//PEAK CALLING//DNASE SEQ |
| 10 | 0.0000085628 | ARID3B//JUMONJI//AT RICH INTERACTION DOMAIN |