Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
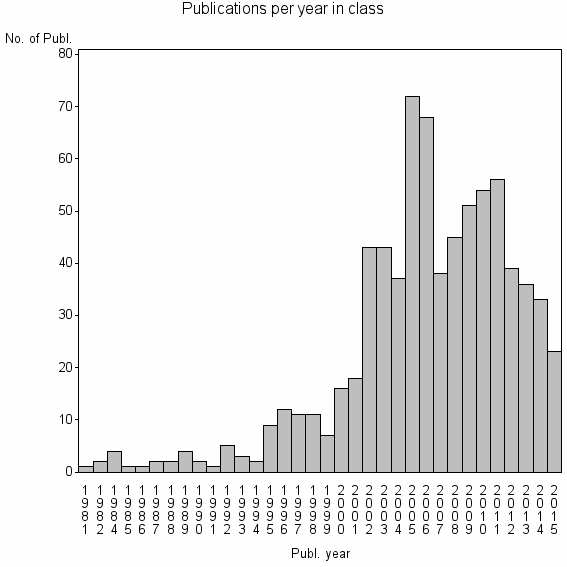
| 13760 | 752 | 25.0 | 58% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | GENOME REARRANGEMENTS | Author keyword | 62 | 42% | 15% | 115 |
| 2 | SORTING BY REVERSALS | Author keyword | 46 | 82% | 4% | 27 |
| 3 | BREAKPOINT GRAPH | Author keyword | 34 | 93% | 2% | 13 |
| 4 | COMMON INTERVALS | Author keyword | 17 | 79% | 1% | 11 |
| 5 | DCJ | Author keyword | 13 | 80% | 1% | 8 |
| 6 | GENOMIC DISTANCE | Author keyword | 12 | 75% | 1% | 9 |
| 7 | GENE TEAMS | Author keyword | 12 | 86% | 1% | 6 |
| 8 | INVERSION DISTANCE | Author keyword | 11 | 100% | 1% | 6 |
| 9 | SYNTENIC DISTANCE | Author keyword | 11 | 100% | 1% | 6 |
| 10 | DATA MIN SEARCH GRP | Address | 9 | 83% | 1% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENOME REARRANGEMENTS | 62 | 42% | 15% | 115 | Search GENOME+REARRANGEMENTS | Search GENOME+REARRANGEMENTS |
| 2 | SORTING BY REVERSALS | 46 | 82% | 4% | 27 | Search SORTING+BY+REVERSALS | Search SORTING+BY+REVERSALS |
| 3 | BREAKPOINT GRAPH | 34 | 93% | 2% | 13 | Search BREAKPOINT+GRAPH | Search BREAKPOINT+GRAPH |
| 4 | COMMON INTERVALS | 17 | 79% | 1% | 11 | Search COMMON+INTERVALS | Search COMMON+INTERVALS |
| 5 | DCJ | 13 | 80% | 1% | 8 | Search DCJ | Search DCJ |
| 6 | GENOMIC DISTANCE | 12 | 75% | 1% | 9 | Search GENOMIC+DISTANCE | Search GENOMIC+DISTANCE |
| 7 | GENE TEAMS | 12 | 86% | 1% | 6 | Search GENE+TEAMS | Search GENE+TEAMS |
| 8 | INVERSION DISTANCE | 11 | 100% | 1% | 6 | Search INVERSION+DISTANCE | Search INVERSION+DISTANCE |
| 9 | SYNTENIC DISTANCE | 11 | 100% | 1% | 6 | Search SYNTENIC+DISTANCE | Search SYNTENIC+DISTANCE |
| 10 | SORTING BY TRANSPOSITIONS | 9 | 67% | 1% | 8 | Search SORTING+BY+TRANSPOSITIONS | Search SORTING+BY+TRANSPOSITIONS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SIGNED PERMUTATIONS | 93 | 79% | 8% | 60 |
| 2 | REVERSALS | 30 | 19% | 19% | 141 |
| 3 | GENOME REARRANGEMENTS | 27 | 31% | 10% | 72 |
| 4 | COMMON INTERVALS | 23 | 60% | 3% | 25 |
| 5 | 15 APPROXIMATION ALGORITHM | 22 | 81% | 2% | 13 |
| 6 | 1375 APPROXIMATION ALGORITHM | 21 | 90% | 1% | 9 |
| 7 | DCJ | 21 | 85% | 1% | 11 |
| 8 | SORTING SIGNED PERMUTATIONS | 21 | 85% | 1% | 11 |
| 9 | GENE ORDERS | 15 | 77% | 1% | 10 |
| 10 | TRANSPOSITIONS | 14 | 30% | 5% | 38 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Algorithmic approaches for genome rearrangement: A review | 2006 | 8 | 37 | 84% |
| Structural dynamics of eukaryotic chromosome evolution | 2003 | 225 | 52 | 13% |
| Ancestral animal genomes reconstruction | 2007 | 11 | 25 | 28% |
| Advances in phylogeny reconstruction from gene order and content data | 2005 | 10 | 44 | 43% |
| Counting on comparative maps | 1998 | 74 | 48 | 29% |
| GENOMIC DIVERGENCE THROUGH GENE REARRANGEMENT | 1990 | 12 | 2 | 50% |
| Large and small rearrangements in the evolution of prokaryotic genomes | 2006 | 0 | 37 | 46% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DATA MIN SEARCH GRP | 9 | 83% | 0.7% | 5 |
| 2 | EPFL IC LCBB | 3 | 100% | 0.4% | 3 |
| 3 | AG GENOMINFORMAT | 3 | 25% | 1.3% | 10 |
| 4 | LACIM | 3 | 13% | 2.5% | 19 |
| 5 | IGM INFO | 2 | 33% | 0.7% | 5 |
| 6 | CGL | 2 | 43% | 0.4% | 3 |
| 7 | EQUIPE EBM | 1 | 100% | 0.3% | 2 |
| 8 | IIS LCBB | 1 | 100% | 0.3% | 2 |
| 9 | PUBL HLTH INFORMAT FELLOW PROGRAM | 1 | 100% | 0.3% | 2 |
| 10 | TEAM BAMBOO | 1 | 100% | 0.3% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000143505 | WHOLE GENOME DUPLICATION//GENET MOL CELLULAIRE ECT LEVU INTERE//GENOME DUPLICATION |
| 2 | 0.0000135503 | CIRCOS//BIODADOS//ORTHOLOGY |
| 3 | 0.0000098949 | MULTIPLE SEQUENCE ALIGNMENT//T COFFEE//STATISTICAL ALIGNMENT |
| 4 | 0.0000094909 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 5 | 0.0000088941 | PLATYRRHINI//NEW WORLD MONKEYS//EVOLUTIONARY BREAKPOINTS |
| 6 | 0.0000085532 | LONGEST COMMON SUBSEQUENCE//CONSTRAINED LONGEST COMMON SUBSEQUENCE//SHORTEST COMMON SUPERSTRING |
| 7 | 0.0000084759 | BIDIRECTIONAL PROMOTER//NEIGHBORING GENES//GENOME NETWORK PROJECT CORE GRPGENOM SCI |
| 8 | 0.0000057580 | TREE OF LIFE//HORIZONTAL GENE TRANSFER//LATERAL GENE TRANSFER |
| 9 | 0.0000054750 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 10 | 0.0000054644 | IROQUOIS//CONSERVED NONCODING ELEMENTS//ENERGY ENVIRONM BIOL COMP |