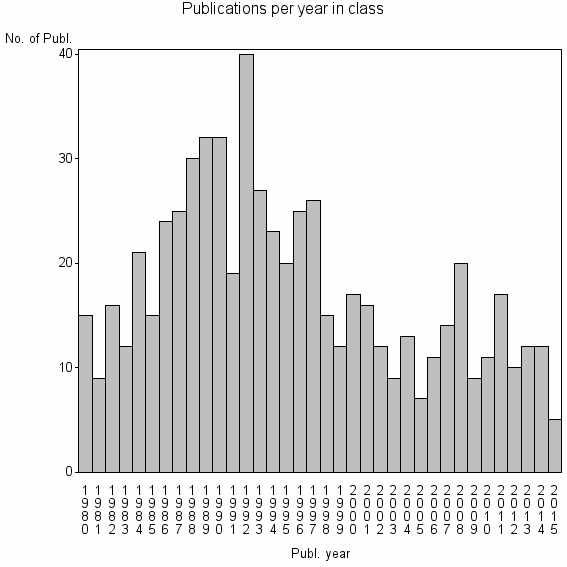
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 15604 | 633 | 34.2 | 60% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 587 | 13504 | NEW ZEALAND FLORA//NEW ZEALAND JOURNAL OF BOTANY//GENOME SIZE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RDNA INTERGENIC SPACER | Author keyword | 11 | 78% | 1% | 7 |
| 2 | NON CONCERTED EVOLUTION | Author keyword | 6 | 80% | 1% | 4 |
| 3 | INTERGENIC SPACER | Author keyword | 4 | 17% | 4% | 23 |
| 4 | NOR EXPRESSION | Author keyword | 4 | 75% | 0% | 3 |
| 5 | AESCHYNANTHUS | Author keyword | 3 | 57% | 1% | 4 |
| 6 | BIPARTITION NETWORKS | Author keyword | 3 | 100% | 0% | 3 |
| 7 | NUCLEOLAR DOMINANCE | Author keyword | 3 | 26% | 1% | 9 |
| 8 | RDNA EXPRESSION | Author keyword | 3 | 60% | 0% | 3 |
| 9 | NOR LOCI | Author keyword | 2 | 67% | 0% | 2 |
| 10 | SPACER LENGTH VARIANTS | Author keyword | 2 | 50% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RDNA INTERGENIC SPACER | 11 | 78% | 1% | 7 | Search RDNA+INTERGENIC+SPACER | Search RDNA+INTERGENIC+SPACER |
| 2 | NON CONCERTED EVOLUTION | 6 | 80% | 1% | 4 | Search NON+CONCERTED+EVOLUTION | Search NON+CONCERTED+EVOLUTION |
| 3 | INTERGENIC SPACER | 4 | 17% | 4% | 23 | Search INTERGENIC+SPACER | Search INTERGENIC+SPACER |
| 4 | NOR EXPRESSION | 4 | 75% | 0% | 3 | Search NOR+EXPRESSION | Search NOR+EXPRESSION |
| 5 | AESCHYNANTHUS | 3 | 57% | 1% | 4 | Search AESCHYNANTHUS | Search AESCHYNANTHUS |
| 6 | BIPARTITION NETWORKS | 3 | 100% | 0% | 3 | Search BIPARTITION+NETWORKS | Search BIPARTITION+NETWORKS |
| 7 | NUCLEOLAR DOMINANCE | 3 | 26% | 1% | 9 | Search NUCLEOLAR+DOMINANCE | Search NUCLEOLAR+DOMINANCE |
| 8 | RDNA EXPRESSION | 3 | 60% | 0% | 3 | Search RDNA+EXPRESSION | Search RDNA+EXPRESSION |
| 9 | NOR LOCI | 2 | 67% | 0% | 2 | Search NOR+LOCI | Search NOR+LOCI |
| 10 | SPACER LENGTH VARIANTS | 2 | 50% | 0% | 3 | Search SPACER+LENGTH+VARIANTS | Search SPACER+LENGTH+VARIANTS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EXTERNAL SPACER | 32 | 80% | 3% | 20 |
| 2 | SILVER STAINING PROCEDURE | 13 | 80% | 1% | 8 |
| 3 | INTERGENIC SPACER | 8 | 16% | 8% | 49 |
| 4 | 17S 25S SPACER REGION | 6 | 80% | 1% | 4 |
| 5 | RDNA INTERGENIC SPACER | 5 | 42% | 2% | 10 |
| 6 | LENGTH VARIANTS | 4 | 41% | 1% | 7 |
| 7 | X RYE HYBRIDS | 3 | 50% | 1% | 5 |
| 8 | DNA SPACER REGION | 3 | 100% | 0% | 3 |
| 9 | P MARIANA | 3 | 100% | 0% | 3 |
| 10 | SERPENTINE ENDEMICS | 3 | 100% | 0% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants | 2007 | 211 | 69 | 26% |
| RIBOSOMAL-RNA GENES IN PLANTS - VARIABILITY IN COPY NUMBER AND IN THE INTERGENIC SPACER | 1987 | 342 | 71 | 62% |
| Ribosomal ITS sequences and plant phylogenetic inference | 2003 | 886 | 153 | 18% |
| Nucleolar dominance and silencing of transcription | 1999 | 70 | 31 | 42% |
| The epigenetics of nucleolar dominance | 2000 | 95 | 27 | 30% |
| Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids | 2004 | 46 | 78 | 36% |
| Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression | 2007 | 0 | 137 | 33% |
| ASSAYING DIFFERENTIAL RIBOSOMAL-RNA GENE-EXPRESSION WITH ALLELE-SPECIFIC PROBES | 1993 | 1 | 14 | 21% |
| NUCLEAR-DNA MARKERS IN ANGIOSPERM TAXONOMY | 1992 | 6 | 98 | 16% |
| REPETITIVITY AND VARIABILITY OF HIGHER-PLANT GENOMES | 1987 | 28 | 116 | 15% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOGEOL PL PALEONTOL | 1 | 50% | 0.2% | 1 |
| 2 | BOT LICADA AGRICULTURA | 1 | 50% | 0.2% | 1 |
| 3 | GEN GENT | 1 | 50% | 0.2% | 1 |
| 4 | GENOM BIOTECHNOL CGB | 1 | 50% | 0.2% | 1 |
| 5 | LBVPAM | 1 | 50% | 0.2% | 1 |
| 6 | MARINE SCI TROP BIOL | 1 | 50% | 0.2% | 1 |
| 7 | MOLEC LIFE SCI | 1 | 50% | 0.2% | 1 |
| 8 | SECCAO GENET | 0 | 13% | 0.5% | 3 |
| 9 | CNRS IRD U V | 0 | 33% | 0.2% | 1 |
| 10 | CONSERVATOIRE JARDIN BOT | 0 | 33% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000296993 | PACHYTENE CHROMOSOME//FLUOROCHROME BANDING//PACHYTENE CHROMOSOMES |
| 2 | 0.0000224915 | MOL BIOL CELL 2//RNA POLYMERASE I//UBF |
| 3 | 0.0000161756 | MOLECULAR AUTHENTICATION//NRDNA ITS REGION//ZHEJIANG PROV GENET IMPROVEMENT QUAL CO |
| 4 | 0.0000127284 | PAEONIA//INVERSION HETEROZYGOSITY//PAEONIA LUDLOWII |
| 5 | 0.0000123326 | POKEY//LAMPBRUSH LOOPS//FERTILITY GENES |
| 6 | 0.0000122805 | NARCISSUS SPP//MOSQUITO CELL LINES//PLANT CELL NUCLEUS |
| 7 | 0.0000116505 | POLYPLOIDY//AUTOPOLYPLOIDY//ALLOPOLYPLOIDY |
| 8 | 0.0000102162 | ARMERIA//ARMERIA MARITIMA//ARMERIA CAESPITOSA |
| 9 | 0.0000096861 | TRITICEAE//ELYMUS//LEYMUS |
| 10 | 0.0000096801 | JUAN FERNANDEZ ISLANDS//DUBAUTIA//TARWEEDS |