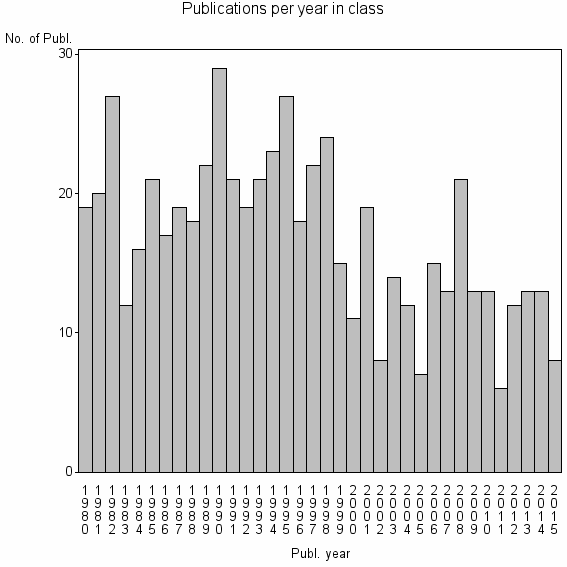
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 16068 | 608 | 38.4 | 59% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1138 | 9139 | HYDROPHOBIN//FUNGAL GENETICS AND BIOLOGY//USTILAGO MAYDIS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | NITROGEN METABOLITE REPRESSION | Author keyword | 21 | 73% | 3% | 16 |
| 2 | NMRA | Author keyword | 6 | 71% | 1% | 5 |
| 3 | FACB | Author keyword | 6 | 100% | 1% | 4 |
| 4 | NIT2 | Author keyword | 3 | 50% | 1% | 5 |
| 5 | MEAB | Author keyword | 3 | 60% | 0% | 3 |
| 6 | PLANT SCI FUNGAL MOL BIOL GRP | Address | 3 | 60% | 0% | 3 |
| 7 | UMR CNRS C8621 | Address | 3 | 60% | 0% | 3 |
| 8 | GATA PROTEIN | Author keyword | 2 | 67% | 0% | 2 |
| 9 | NIT4 | Author keyword | 2 | 67% | 0% | 2 |
| 10 | NMR1 | Author keyword | 2 | 67% | 0% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NITROGEN METABOLITE REPRESSION | 21 | 73% | 3% | 16 | Search NITROGEN+METABOLITE+REPRESSION | Search NITROGEN+METABOLITE+REPRESSION |
| 2 | NMRA | 6 | 71% | 1% | 5 | Search NMRA | Search NMRA |
| 3 | FACB | 6 | 100% | 1% | 4 | Search FACB | Search FACB |
| 4 | NIT2 | 3 | 50% | 1% | 5 | Search NIT2 | Search NIT2 |
| 5 | MEAB | 3 | 60% | 0% | 3 | Search MEAB | Search MEAB |
| 6 | GATA PROTEIN | 2 | 67% | 0% | 2 | Search GATA+PROTEIN | Search GATA+PROTEIN |
| 7 | NIT4 | 2 | 67% | 0% | 2 | Search NIT4 | Search NIT4 |
| 8 | NMR1 | 2 | 67% | 0% | 2 | Search NMR1 | Search NMR1 |
| 9 | UAPC | 2 | 67% | 0% | 2 | Search UAPC | Search UAPC |
| 10 | AREB | 1 | 38% | 0% | 3 | Search AREB | Search AREB |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ACID XANTHINE PERMEASE | 72 | 93% | 4% | 27 |
| 2 | NITROGEN METABOLITE REPRESSION | 71 | 61% | 12% | 75 |
| 3 | METABOLITE REPRESSION | 40 | 71% | 5% | 32 |
| 4 | XANTHINE PERMEASE | 34 | 93% | 2% | 13 |
| 5 | GATA FACTOR AREA | 34 | 63% | 6% | 34 |
| 6 | CYSTEINE SCANNING ANALYSIS | 31 | 92% | 2% | 12 |
| 7 | NAT NCS2 FAMILY | 30 | 100% | 2% | 12 |
| 8 | PUTATIVE ZINC FINGER | 25 | 43% | 7% | 44 |
| 9 | INITIATOR CONSTITUTIVE MUTATION | 24 | 91% | 2% | 10 |
| 10 | PURINE TRANSPORTERS | 23 | 79% | 2% | 15 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Genetic regulation of nitrogen metabolism in the fungi | 1997 | 372 | 98 | 52% |
| Nitrogen regulation of fungal secondary metabolism in fungi | 2014 | 4 | 117 | 52% |
| REGULATION OF SULFUR AND NITROGEN-METABOLISM IN FILAMENTOUS FUNGI | 1993 | 75 | 50 | 58% |
| Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi | 2008 | 44 | 112 | 31% |
| Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the "streetwise" GATA family of transcription factors | 1998 | 67 | 69 | 45% |
| Fungal nucleobase transporters | 2007 | 15 | 72 | 39% |
| The fungal GATA factors | 2000 | 104 | 48 | 27% |
| NITROGEN CATABOLITE REPRESSION IN YEASTS AND FILAMENTOUS FUNGI | 1985 | 183 | 69 | 43% |
| The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role | 2008 | 19 | 90 | 34% |
| The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi | 2008 | 18 | 87 | 31% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT SCI FUNGAL MOL BIOL GRP | 3 | 60% | 0.5% | 3 |
| 2 | UMR CNRS C8621 | 3 | 60% | 0.5% | 3 |
| 3 | UMR C8621 | 2 | 26% | 0.8% | 5 |
| 4 | AUSTRIAN TECHNOL GMBH AIT | 1 | 100% | 0.3% | 2 |
| 5 | INVEST BIOMED BIOTECNOL | 1 | 50% | 0.3% | 2 |
| 6 | FUNGAL GENET GENOM UNIT | 1 | 19% | 0.8% | 5 |
| 7 | GENET MICROBICA | 1 | 33% | 0.3% | 2 |
| 8 | FUNGAL GENOM UNIT | 1 | 23% | 0.5% | 3 |
| 9 | BOT WESTFAL | 1 | 50% | 0.2% | 1 |
| 10 | INTERDISCIPLINAIRE ENVIRONM CONTINENTAUXUMR | 1 | 50% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000268686 | RSP5 UBIQUITIN LIGASE//GLN3//RSP5 |
| 2 | 0.0000203980 | L GLUTAMINASE//MICROCOCCUS LUTEUS K 3//GLUTAMINASE |
| 3 | 0.0000192782 | HYGROMYCIN B RESISTANCE//FUNGAL TRANSFORMATION//ATMT |
| 4 | 0.0000178566 | XLNR//AMYR//TRICHODERMA REESEI |
| 5 | 0.0000156268 | PACC//PI SENSING//HUMICOLA LUTEA |
| 6 | 0.0000125593 | GAL4P//GAL80//GAL GENES |
| 7 | 0.0000123848 | VEA//LAEA//ASPERGILLUS NIDULANS |
| 8 | 0.0000097733 | NITRATE REDUCTASE//NITRITE REDUCTASE//NADPH NITRATE REDUCTASE |
| 9 | 0.0000097563 | SORDARIA MACROSPORA//LEHRSTUHL ALLGEMEINE MOL BOT//PSEUDOHOMOTHALLISM |
| 10 | 0.0000069407 | SULFUR REGULATION//HOMOCYSTEINE SYNTHASE//CYS3 |