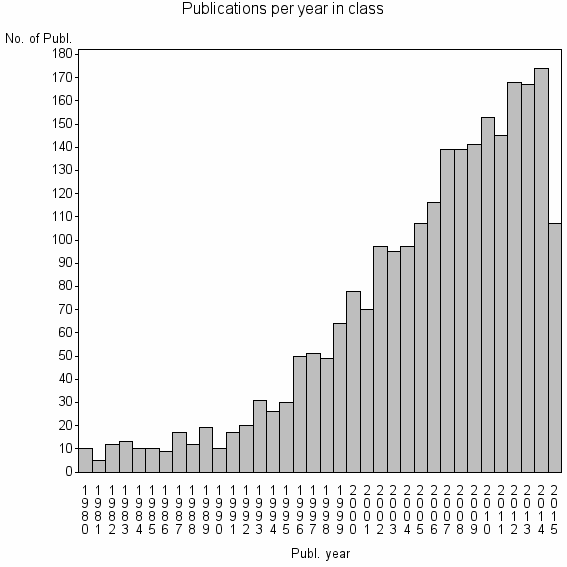
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 1680 | 2458 | 49.6 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1846 | 5606 | LANDSCAPE GENETICS//INBREEDING DEPRESSION//COALESCENT |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MCDONALD KREITMAN TEST | Author keyword | 38 | 76% | 1% | 26 |
| 2 | COALESCENT | Author keyword | 33 | 28% | 4% | 101 |
| 3 | SELECTIVE SWEEPS | Author keyword | 23 | 52% | 1% | 32 |
| 4 | STRUCTURED COALESCENT | Author keyword | 22 | 81% | 1% | 13 |
| 5 | ANCESTRAL INFERENCE | Author keyword | 19 | 80% | 0% | 12 |
| 6 | ANCESTRAL SELECTION GRAPH | Author keyword | 19 | 80% | 0% | 12 |
| 7 | SITE FREQUENCY SPECTRUM | Author keyword | 17 | 70% | 1% | 14 |
| 8 | SELECTIVE SWEEP | Author keyword | 14 | 30% | 2% | 41 |
| 9 | GENETIC HITCHHIKING | Author keyword | 14 | 48% | 1% | 22 |
| 10 | ALLELE FREQUENCY SPECTRUM | Author keyword | 14 | 100% | 0% | 7 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MCDONALD KREITMAN TEST | 38 | 76% | 1% | 26 | Search MCDONALD+KREITMAN+TEST | Search MCDONALD+KREITMAN+TEST |
| 2 | COALESCENT | 33 | 28% | 4% | 101 | Search COALESCENT | Search COALESCENT |
| 3 | SELECTIVE SWEEPS | 23 | 52% | 1% | 32 | Search SELECTIVE+SWEEPS | Search SELECTIVE+SWEEPS |
| 4 | STRUCTURED COALESCENT | 22 | 81% | 1% | 13 | Search STRUCTURED+COALESCENT | Search STRUCTURED+COALESCENT |
| 5 | ANCESTRAL INFERENCE | 19 | 80% | 0% | 12 | Search ANCESTRAL+INFERENCE | Search ANCESTRAL+INFERENCE |
| 6 | ANCESTRAL SELECTION GRAPH | 19 | 80% | 0% | 12 | Search ANCESTRAL+SELECTION+GRAPH | Search ANCESTRAL+SELECTION+GRAPH |
| 7 | SITE FREQUENCY SPECTRUM | 17 | 70% | 1% | 14 | Search SITE+FREQUENCY+SPECTRUM | Search SITE+FREQUENCY+SPECTRUM |
| 8 | SELECTIVE SWEEP | 14 | 30% | 2% | 41 | Search SELECTIVE+SWEEP | Search SELECTIVE+SWEEP |
| 9 | GENETIC HITCHHIKING | 14 | 48% | 1% | 22 | Search GENETIC+HITCHHIKING | Search GENETIC+HITCHHIKING |
| 10 | ALLELE FREQUENCY SPECTRUM | 14 | 100% | 0% | 7 | Search ALLELE+FREQUENCY+SPECTRUM | Search ALLELE+FREQUENCY+SPECTRUM |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BACKGROUND SELECTION | 171 | 62% | 7% | 179 |
| 2 | SEGREGATING SITES | 127 | 65% | 5% | 122 |
| 3 | HITCHHIKING | 108 | 53% | 6% | 143 |
| 4 | NEUTRAL MOLECULAR VARIATION | 83 | 69% | 3% | 71 |
| 5 | ADAPTIVE PROTEIN EVOLUTION | 68 | 51% | 4% | 96 |
| 6 | RECENT POSITIVE SELECTION | 61 | 41% | 5% | 114 |
| 7 | DNA POLYMORPHISM DATA | 54 | 60% | 2% | 59 |
| 8 | COALESCENT PROCESS | 52 | 64% | 2% | 51 |
| 9 | MOLECULAR POPULATION GENETICS | 50 | 36% | 5% | 112 |
| 10 | GENEALOGICAL PROCESS | 47 | 88% | 1% | 22 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Combining experimental evolution with next-generation sequencing: a powerful tool to study adaptation from standing genetic variation | 2015 | 5 | 41 | 49% |
| Effective population size and patterns of molecular evolution and variation | 2009 | 304 | 107 | 60% |
| Adaptation from standing genetic variation | 2008 | 421 | 64 | 50% |
| The Genetics of Human Adaptation: Hard Sweeps, Soft Sweeps, and Polygenic Adaptation | 2010 | 220 | 68 | 49% |
| Genomic signatures of selection at linked sites: unifying the disparity among species | 2013 | 45 | 155 | 66% |
| Constructing genomic maps of positive selection in humans: Where do we go from here? | 2009 | 179 | 94 | 61% |
| FUNDAMENTAL CONCEPTS IN GENETICS Genetics in geographically structured populations: defining, estimating and interpreting F-ST | 2009 | 271 | 70 | 41% |
| Molecular signatures of natural selection | 2005 | 598 | 114 | 54% |
| Pervasive Natural Selection in the Drosophila Genome? | 2009 | 129 | 115 | 77% |
| Positive natural selection in the human lineage | 2006 | 470 | 60 | 47% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SECT EVOLUTIONARY BIOL | 12 | 28% | 1.5% | 37 |
| 2 | BIOMED SCI TECHNOL LITA SEGRATE | 7 | 64% | 0.3% | 7 |
| 3 | BIOL STAT COMPUTAT BIOL | 5 | 11% | 1.7% | 43 |
| 4 | BIOL S 2102 | 4 | 67% | 0.2% | 4 |
| 5 | BIOL S 288 | 3 | 100% | 0.1% | 3 |
| 6 | STUDY EVOLUT | 3 | 12% | 0.9% | 21 |
| 7 | FDN CA GRANDA IRCCS OSPED MAGGIORE POLICLIN | 3 | 30% | 0.3% | 7 |
| 8 | UNIT HUMAN EVOLUTIONARY GENET | 2 | 26% | 0.3% | 8 |
| 9 | GRP GENOM BIOINFORMAT EVOLUT | 2 | 67% | 0.1% | 2 |
| 10 | MANGIAGALLI REGINA ELENA FDNDINO FERRARI | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000219224 | BRITONS//BELGAE//BIOL S 4100 |
| 2 | 0.0000151239 | TEMPORAL METHOD//EFFECTIVE POPULATION SIZE//N E N RATIO |
| 3 | 0.0000151078 | APPROXIMATE BAYESIAN COMPUTATION//LIKELIHOOD FREE INFERENCE//LIKELIHOOD FREE METHODS |
| 4 | 0.0000129644 | ISOCHORES//EVOLUZ MOL//ISOCHORE |
| 5 | 0.0000107595 | SEX BIASED GENE EXPRESSION//SEXUAL ANTAGONISM//INTRALOCUS SEXUAL CONFLICT |
| 6 | 0.0000104989 | ASCOBOLUS//EVOLUTION OF PLOIDY//MULTIGENE ARRAYS |
| 7 | 0.0000098395 | MOLECULAR CLOCK//RELAXED CLOCK//PHYLODYNAMICS |
| 8 | 0.0000097676 | OPSARIICHTHYS//NESTED CLADE ANALYSIS//POPULATION EXPANSION |
| 9 | 0.0000097146 | HAPLOTYPE INFERENCE//PURE PARSIMONY//HAPLOTYPE ASSEMBLY |
| 10 | 0.0000093583 | MAPPING BY SEQUENCING//RADSEQ//REDUCED REPRESENTATION LIBRARIES |