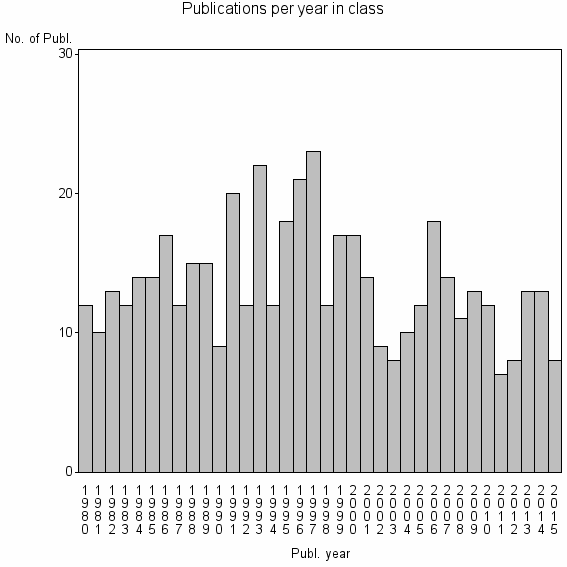
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 18410 | 487 | 33.7 | 69% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 302 | 17871 | TRANSLESION SYNTHESIS//DNA POLYMERASE//RECA PROTEIN |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EMANUEL SYNDROME | Author keyword | 11 | 78% | 1% | 7 |
| 2 | PARTIAL TRISOMY 11Q | Author keyword | 9 | 83% | 1% | 5 |
| 3 | DUPLICATION 11Q | Author keyword | 8 | 100% | 1% | 5 |
| 4 | DER22 | Author keyword | 4 | 75% | 1% | 3 |
| 5 | SISTER CHROMOSOME EXCHANGE | Author keyword | 4 | 75% | 1% | 3 |
| 6 | PATRR | Author keyword | 3 | 100% | 1% | 3 |
| 7 | QUASIPALINDROME | Author keyword | 3 | 100% | 1% | 3 |
| 8 | 11 22 TRANSLOCATION | Author keyword | 2 | 67% | 0% | 2 |
| 9 | PALINDROME | Author keyword | 2 | 11% | 3% | 17 |
| 10 | DIRECT REPEATS | Author keyword | 2 | 18% | 2% | 10 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | EMANUEL SYNDROME | 11 | 78% | 1% | 7 | Search EMANUEL+SYNDROME | Search EMANUEL+SYNDROME |
| 2 | PARTIAL TRISOMY 11Q | 9 | 83% | 1% | 5 | Search PARTIAL+TRISOMY+11Q | Search PARTIAL+TRISOMY+11Q |
| 3 | DUPLICATION 11Q | 8 | 100% | 1% | 5 | Search DUPLICATION+11Q | Search DUPLICATION+11Q |
| 4 | DER22 | 4 | 75% | 1% | 3 | Search DER22 | Search DER22 |
| 5 | SISTER CHROMOSOME EXCHANGE | 4 | 75% | 1% | 3 | Search SISTER+CHROMOSOME+EXCHANGE | Search SISTER+CHROMOSOME+EXCHANGE |
| 6 | PATRR | 3 | 100% | 1% | 3 | Search PATRR | Search PATRR |
| 7 | QUASIPALINDROME | 3 | 100% | 1% | 3 | Search QUASIPALINDROME | Search QUASIPALINDROME |
| 8 | 11 22 TRANSLOCATION | 2 | 67% | 0% | 2 | Search 11+22+TRANSLOCATION | Search 11+22+TRANSLOCATION |
| 9 | PALINDROME | 2 | 11% | 3% | 17 | Search PALINDROME | Search PALINDROME |
| 10 | DIRECT REPEATS | 2 | 18% | 2% | 10 | Search DIRECT+REPEATS | Search DIRECT+REPEATS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | 22Q11 BREAKPOINTS | 31 | 92% | 2% | 12 |
| 2 | CLUSTERED 11Q23 | 23 | 86% | 2% | 12 |
| 3 | RECURRENT CONSTITUTIONAL T11 22 | 23 | 72% | 4% | 18 |
| 4 | ESCHERICHIA COLI PLASMIDS | 16 | 58% | 4% | 19 |
| 5 | DNA PALINDROMES | 14 | 59% | 3% | 16 |
| 6 | AT RICH PALINDROMES | 14 | 46% | 5% | 23 |
| 7 | 1ST MEIOSIS | 11 | 78% | 1% | 7 |
| 8 | BIO TRANSDUCING PHAGE | 10 | 63% | 2% | 10 |
| 9 | 11Q 22Q TRANSLOCATION | 10 | 44% | 3% | 17 |
| 10 | DELETION FORMATION | 10 | 33% | 5% | 24 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Prevalence of Emanuel syndrome: Theoretical frequency and surveillance result | 2014 | 1 | 20 | 100% |
| The constitutional t(11;22): implications for a novel mechanism responsible for gross chromosomal rearrangements | 2010 | 17 | 71 | 56% |
| Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences | 2004 | 126 | 71 | 46% |
| Chromosomal translocations mediated by palindromic DNA | 2006 | 17 | 44 | 61% |
| Anesthesia in a patient with chromosome 11;22 translocation: a case report and literature review | 2005 | 3 | 6 | 100% |
| Phenotype and Micro-array characterization of duplication 11q22.1-q25 and review of the literature | 2013 | 3 | 22 | 55% |
| LONG DNA PALINDROMES, CRUCIFORM STRUCTURES, GENETIC INSTABILITY AND SECONDARY STRUCTURE REPAIR | 1994 | 179 | 50 | 36% |
| Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells | 2007 | 47 | 104 | 21% |
| A new case of de novo 11q duplication in a patient with normal development and intelligence and review of the literature | 2007 | 13 | 14 | 43% |
| A NOVEL ASSAY FOR ILLEGITIMATE RECOMBINATION IN ESCHERICHIA-COLI - STIMULATION OF LAMBDA-BIO TRANSDUCING PHAGE FORMATION BY ULTRAVIOLET-LIGHT AND ITS INDEPENDENCE FROM RECA FUNCTION | 1995 | 16 | 26 | 65% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DEV TARGETED MINIMALLY INVAS DIAG TREATME | 2 | 28% | 1.0% | 5 |
| 2 | ABRAMSON 1002 | 1 | 30% | 0.6% | 3 |
| 3 | BIOSCI TECHNOL BIOCHEM BIOPHYS | 1 | 50% | 0.2% | 1 |
| 4 | GENET METAB CENT | 1 | 50% | 0.2% | 1 |
| 5 | LFS UNIT | 1 | 50% | 0.2% | 1 |
| 6 | POST GEN DIS | 1 | 50% | 0.2% | 1 |
| 7 | SERV NEONATOL GENET | 1 | 50% | 0.2% | 1 |
| 8 | GENOME DNA STRUCT MUTAGENESIS | 1 | 29% | 0.4% | 2 |
| 9 | ROSENSTIEL BASIC MED SCI MS029 | 1 | 25% | 0.4% | 2 |
| 10 | BIOCOMP INFORMAT | 1 | 22% | 0.4% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000174614 | JACOBSEN SYNDROME//11Q SYNDROME//11Q TERMINAL DELETION DISORDER |
| 2 | 0.0000155360 | HOLLIDAY JUNCTION//RECBCD ENZYME//RECBCD |
| 3 | 0.0000150428 | FEN1//BRAUN S//CREAT INITIAT CELL CYCLE CONTROL |
| 4 | 0.0000119498 | EPIDEMIOLOGICAL SIGNIFICANCE//SIMILARITY IDENTIFICATION//AIRBORNE ESCHERICHIA COLI |
| 5 | 0.0000111403 | ROLLING CIRCLE REPLICATION//PLASMID REPLICATION CONTROL//CRYPTIC PLASMID |
| 6 | 0.0000094724 | ADAPTIVE MUTATION//STATIONARY PHASE MUTATION//STATIONARY PHASE MUTAGENESIS |
| 7 | 0.0000092887 | COMPLEX CHROMOSOMAL REARRANGEMENT//COMPLEX CHROMOSOME REARRANGEMENT CCR//COMPLEX CHROMOSOMAL REARRANGEMENTS |
| 8 | 0.0000090002 | CAT EYE SYNDROME//SUPERNUMERARY MARKER CHROMOSOMES//SMALL SUPERNUMERARY MARKER CHROMOSOME |
| 9 | 0.0000078472 | SPI SELECTION//CO PRECIPITANT//INSERT SIZE |
| 10 | 0.0000071343 | PALLISTER KILLIAN SYNDROME//TETRASOMY 12P//TRISOMY 12P |