Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
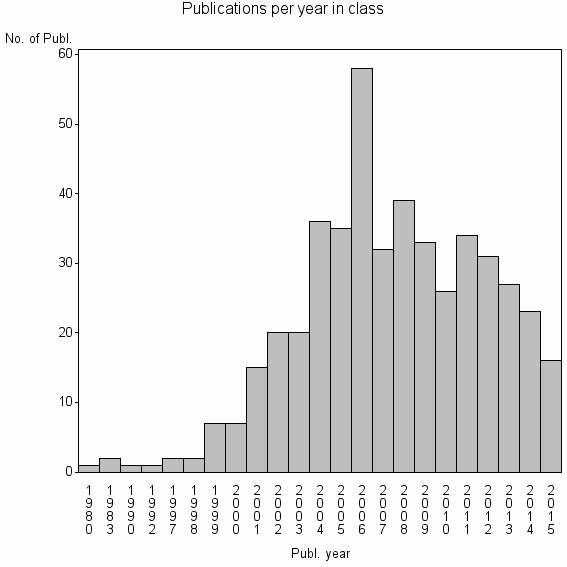
| 18835 | 468 | 53.1 | 88% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 356 | 16803 | PROTEOMICS//JOURNAL OF PROTEOME RESEARCH//PROTEIN IDENTIFICATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SYNGAP1 | Author keyword | 8 | 70% | 1% | 7 |
| 2 | GENES COGNIT PROGRAMME | Address | 5 | 34% | 2% | 11 |
| 3 | NEUROCIENCIAS LIM 27 | Address | 3 | 29% | 2% | 9 |
| 4 | SYNGAP | Author keyword | 2 | 32% | 1% | 6 |
| 5 | DE NOVO SEQUENCE | Author keyword | 2 | 67% | 0% | 2 |
| 6 | GENOM PROTEOM TECHNOL | Address | 2 | 67% | 0% | 2 |
| 7 | GENOMICS TECHNOL | Address | 2 | 67% | 0% | 2 |
| 8 | MOL PHYSIOL SYN SE | Address | 2 | 67% | 0% | 2 |
| 9 | N1E 115 CELL LINE | Author keyword | 2 | 67% | 0% | 2 |
| 10 | PSIQUAITRIA | Address | 2 | 67% | 0% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SYNGAP1 | 8 | 70% | 1% | 7 | Search SYNGAP1 | Search SYNGAP1 |
| 2 | SYNGAP | 2 | 32% | 1% | 6 | Search SYNGAP | Search SYNGAP |
| 3 | DE NOVO SEQUENCE | 2 | 67% | 0% | 2 | Search DE+NOVO+SEQUENCE | Search DE+NOVO+SEQUENCE |
| 4 | N1E 115 CELL LINE | 2 | 67% | 0% | 2 | Search N1E+115+CELL+LINE | Search N1E+115+CELL+LINE |
| 5 | TWO DIMENSIONAL PROTEIN DATABASE | 1 | 50% | 0% | 2 | Search TWO+DIMENSIONAL+PROTEIN+DATABASE | Search TWO+DIMENSIONAL+PROTEIN+DATABASE |
| 6 | MAJOR DEPRESSION DISORDERS | 1 | 40% | 0% | 2 | Search MAJOR+DEPRESSION+DISORDERS | Search MAJOR+DEPRESSION+DISORDERS |
| 7 | PROTEIN MAP | 1 | 20% | 1% | 4 | Search PROTEIN+MAP | Search PROTEIN+MAP |
| 8 | HYPOTHETICAL PROTEINS | 1 | 14% | 1% | 6 | Search HYPOTHETICAL+PROTEINS | Search HYPOTHETICAL+PROTEINS |
| 9 | 2 D LIQUID CHROMATOGRAPHY | 1 | 50% | 0% | 1 | Search 2+D+LIQUID+CHROMATOGRAPHY | Search 2+D+LIQUID+CHROMATOGRAPHY |
| 10 | ARSENITE METHYLTRANSFERASE | 1 | 50% | 0% | 1 | Search ARSENITE+METHYLTRANSFERASE | Search ARSENITE+METHYLTRANSFERASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | POSTSYNAPTIC DENSITY FRACTION | 6 | 21% | 6% | 27 |
| 2 | 2 D DIGE ANALYSIS | 4 | 67% | 1% | 4 |
| 3 | BRAIN PROTEINS | 4 | 19% | 4% | 19 |
| 4 | ENVIRONMENT INTERACTION MODEL | 3 | 60% | 1% | 3 |
| 5 | DOWN SYNDROME BRAIN | 2 | 22% | 1% | 7 |
| 6 | DIHYDROPYRIMIDINASE RELATED PROTEIN 2 | 2 | 15% | 2% | 10 |
| 7 | POSTSYNAPTIC DENSITY 95 | 2 | 13% | 2% | 11 |
| 8 | 2 D DIGE | 1 | 31% | 1% | 4 |
| 9 | MK 801 TREATED RATS | 1 | 38% | 1% | 3 |
| 10 | POSTSYNAPTIC DENSITY FRACTIONS | 1 | 50% | 0% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Synapse proteomics of multiprotein complexes: en route from genes to nervous system diseases | 2005 | 40 | 11 | 45% |
| Evolution of Synapse Complexity and Diversity | 2012 | 19 | 70 | 27% |
| Neuroproteomics: understanding the molecular organization and complexity of the brain | 2009 | 61 | 152 | 26% |
| An Integrated Quantitative Proteomics and Systems Biology Approach to Explore Synaptic Protein Profile Changes During Morphine Exposure | 2014 | 1 | 86 | 35% |
| Maintaining standardization: an update of the HUPO Brain Proteome Project | 2008 | 5 | 25 | 56% |
| Proteomics in brain research: potentials and limitations | 2003 | 95 | 92 | 18% |
| Proteomic research in psychiatry | 2011 | 22 | 195 | 18% |
| The Role of Energy Metabolism Dysfunction and Oxidative Stress in Schizophrenia Revealed by Proteomics | 2011 | 16 | 73 | 19% |
| Recent advances in neuroproteomics | 2007 | 9 | 96 | 34% |
| Proteome analysis of schizophrenia brain tissue | 2010 | 30 | 87 | 15% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENES COGNIT PROGRAMME | 5 | 34% | 2.4% | 11 |
| 2 | NEUROCIENCIAS LIM 27 | 3 | 29% | 1.9% | 9 |
| 3 | GENOM PROTEOM TECHNOL | 2 | 67% | 0.4% | 2 |
| 4 | GENOMICS TECHNOL | 2 | 67% | 0.4% | 2 |
| 5 | MOL PHYSIOL SYN SE | 2 | 67% | 0.4% | 2 |
| 6 | PSIQUAITRIA | 2 | 67% | 0.4% | 2 |
| 7 | GENES COGNIT | 1 | 100% | 0.4% | 2 |
| 8 | MOOD ANXIETY DISORDERS DPU | 1 | 50% | 0.4% | 2 |
| 9 | NEUROPLAST | 1 | 10% | 1.9% | 9 |
| 10 | DISCOVERY MED MOL SCI | 1 | 50% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000199106 | SAMPLE PREFRACTIONATION//PREFRACTIONATION//ALKALINE PROTEINS |
| 2 | 0.0000185834 | MAZUMDAR SHAW TRANSLAT//2 DIMENSIONAL LIQUID CHROMATOGRAPHY//BIOLOGICAL VARIATION ANALYSIS |
| 3 | 0.0000148296 | COLLAPSIN RESPONSE MEDIATOR PROTEIN//CRMP2//CRMP 2 |
| 4 | 0.0000141541 | 2 DE MAP//DATA INDEPENDENT SCANNING//LEUKAEMIA FUND PROTEOM IL |
| 5 | 0.0000101136 | TWO DIMENSIONAL POLYACRYLAMIDE GEL ELECTROPHORESIS//MYOCARDIAL PROTEINS//PROTEOME GENE PROD M PING |
| 6 | 0.0000093216 | BRAIN BANK//AGONAL STATE//BRAIN BANKS |
| 7 | 0.0000091670 | STARGAZIN//PICK1//AMPA RECEPTOR |
| 8 | 0.0000079333 | PROGRAM STRUCT MOL NEUROSCI//STRUCT NEUROSCI//PERFORANT PATH DENTATE GYRUS PATHWAY |
| 9 | 0.0000077917 | N ACETYL ASPARTATE//N ACETYLASPARTATE//MOL DYNAM MENTAL DISORDERS |
| 10 | 0.0000077732 | AMNIOTIC FLUID SUPERNATANT//CERVICAL VAGINAL FLUID//UNITE MED MATERNO FOETALE |