Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
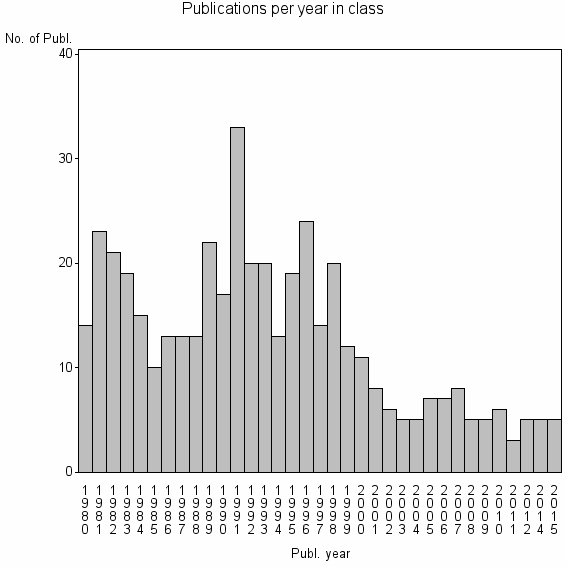
| 19310 | 446 | 40.2 | 60% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 134 | 22710 | SACCHAROMYCES CEREVISIAE//YEAST//SCHIZOSACCHAROMYCES POMBE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEINASE A PRA | Author keyword | 6 | 80% | 1% | 4 |
| 2 | PROTEINASE A | Author keyword | 6 | 42% | 2% | 11 |
| 3 | CARBOXYPEPTIDASE Y | Author keyword | 3 | 22% | 3% | 14 |
| 4 | INDUSTRIAL BREWERS YEAST | Author keyword | 2 | 67% | 0% | 2 |
| 5 | VPS MUTANTS | Author keyword | 2 | 67% | 0% | 2 |
| 6 | PEP4 | Author keyword | 1 | 31% | 1% | 4 |
| 7 | KERATIN LIGAND | Author keyword | 1 | 100% | 0% | 2 |
| 8 | PEP4 GENE | Author keyword | 1 | 100% | 0% | 2 |
| 9 | PHENOTYPIC LAG | Author keyword | 1 | 50% | 0% | 2 |
| 10 | PROTEASE B | Author keyword | 1 | 50% | 0% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEINASE A PRA | 6 | 80% | 1% | 4 | Search PROTEINASE+A+PRA | Search PROTEINASE+A+PRA |
| 2 | PROTEINASE A | 6 | 42% | 2% | 11 | Search PROTEINASE+A | Search PROTEINASE+A |
| 3 | CARBOXYPEPTIDASE Y | 3 | 22% | 3% | 14 | Search CARBOXYPEPTIDASE+Y | Search CARBOXYPEPTIDASE+Y |
| 4 | INDUSTRIAL BREWERS YEAST | 2 | 67% | 0% | 2 | Search INDUSTRIAL+BREWERS+YEAST | Search INDUSTRIAL+BREWERS+YEAST |
| 5 | VPS MUTANTS | 2 | 67% | 0% | 2 | Search VPS+MUTANTS | Search VPS+MUTANTS |
| 6 | PEP4 | 1 | 31% | 1% | 4 | Search PEP4 | Search PEP4 |
| 7 | KERATIN LIGAND | 1 | 100% | 0% | 2 | Search KERATIN+LIGAND | Search KERATIN+LIGAND |
| 8 | PEP4 GENE | 1 | 100% | 0% | 2 | Search PEP4+GENE | Search PEP4+GENE |
| 9 | PHENOTYPIC LAG | 1 | 50% | 0% | 2 | Search PHENOTYPIC+LAG | Search PHENOTYPIC+LAG |
| 10 | PROTEASE B | 1 | 50% | 0% | 2 | Search PROTEASE+B | Search PROTEASE+B |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PEP4 GENE | 22 | 43% | 9% | 39 |
| 2 | HEAD RETENTION | 21 | 90% | 2% | 9 |
| 3 | FOAM PROTEINS | 17 | 100% | 2% | 8 |
| 4 | PROTEINASE YSCB | 14 | 100% | 2% | 7 |
| 5 | PROTEINASE A | 11 | 60% | 3% | 12 |
| 6 | CARBOXYPEPTIDASE Y | 9 | 17% | 11% | 47 |
| 7 | YEAST PROTEINASE | 6 | 80% | 1% | 4 |
| 8 | INVIVO BIOSYNTHESIS | 4 | 47% | 2% | 7 |
| 9 | PROTEASE B | 3 | 33% | 2% | 8 |
| 10 | VACUOLAR HYDROLASES | 3 | 19% | 3% | 12 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| TACKLING THE PROTEASE PROBLEM IN SACCHAROMYCES-CEREVISIAE | 1991 | 320 | 37 | 70% |
| Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae | 2002 | 54 | 36 | 86% |
| Biosynthesis and function of yeast vacuolar proteases - Review | 1996 | 106 | 95 | 68% |
| The structure and function of Saccharomyces cerevisiae proteinase A | 2007 | 33 | 56 | 38% |
| 3 PROTEOLYTIC SYSTEMS IN THE YEAST SACCHAROMYCES-CEREVISIAE | 1991 | 183 | 60 | 48% |
| Vacuole biogenesis in Saccharomyces cerevisiae: Protein transport pathways to the yeast vacuole | 1998 | 188 | 182 | 24% |
| THE FUNGAL VACUOLE - COMPOSITION, FUNCTION, AND BIOGENESIS | 1990 | 501 | 157 | 31% |
| AMINOPEPTIDASE I ENZYMATIC ACTIVITY | 2008 | 4 | 18 | 44% |
| Yeast vacuole inheritance and dynamics | 2003 | 66 | 129 | 23% |
| Receptor-mediated protein sorting to the vacuole in yeast: Roles for a protein kinase, a lipid kinase and GTP-binding proteins | 1995 | 143 | 115 | 23% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOCHEM HERMANN HERDER STR 7 | 1 | 50% | 0.2% | 1 |
| 2 | BIOL MOL GENET MOL DEV | 1 | 50% | 0.2% | 1 |
| 3 | BIOMED S SHINAGAWA KU | 1 | 50% | 0.2% | 1 |
| 4 | MUSASHINO BREWERY | 1 | 50% | 0.2% | 1 |
| 5 | PROTOGENIA | 1 | 50% | 0.2% | 1 |
| 6 | MOL GENET BREEDING YEAST | 0 | 20% | 0.4% | 2 |
| 7 | 258 HIROMACHI 1CHOME | 0 | 25% | 0.2% | 1 |
| 8 | CAROLYN HOFF LYNCH BIOL | 0 | 17% | 0.2% | 1 |
| 9 | GRP MICROBIOL CELULAR LICADA | 0 | 13% | 0.2% | 1 |
| 10 | VIROL BIOL MOL | 0 | 13% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000120876 | PHARM YAMASHIRO CHO//TRYPSIN LIKE PROTEINASE//INTRACELLULAR SERINE PROTEASE |
| 2 | 0.0000112610 | RETROMER//SORTING NEXIN//VPS35 |
| 3 | 0.0000106845 | P I TRANSPORTER//PHO81//PHO89 |
| 4 | 0.0000093509 | LEGUMAIN//ASPARAGINYL ENDOPEPTIDASE//STORAGE PROTEIN MOBILIZATION |
| 5 | 0.0000087630 | PROLYL AMINOPEPTIDASE//TRIPEPTIDYL PEPTIDASE II//AMINOPEPTIDASE |
| 6 | 0.0000086577 | SECRETED ASPARTYL PROTEINASE//SECRETED ASPARTIC PROTEASE//SAP2 |
| 7 | 0.0000085107 | ESCRT//ALG 2//TSG101 |
| 8 | 0.0000083957 | 2 MU M PLASMID//SUC2 PROMOTER//PLASMID STABILITY |
| 9 | 0.0000077663 | SNARE//SYNTAXIN//SYNAPTOTAGMIN |
| 10 | 0.0000076464 | V ATPASE//VACUOLAR ATPASE//V 1 ATPASE |