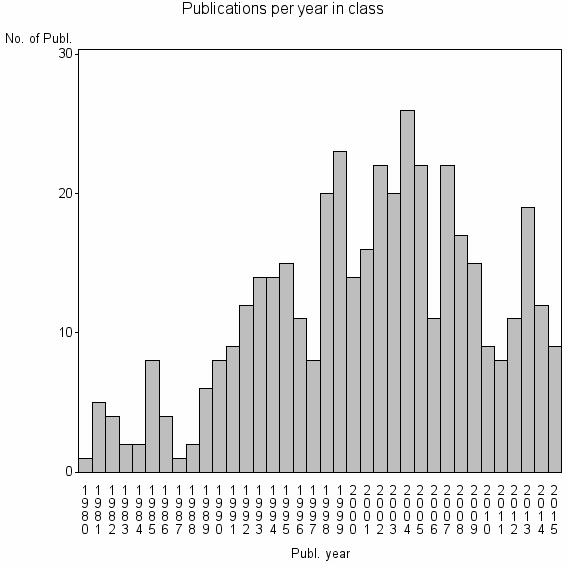
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 19876 | 422 | 40.0 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2279 | 4195 | LIPOYL DOMAIN//PYRUVATE DEHYDROGENASE COMPLEX//NONKETOTIC HYPERGLYCINEMIA |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | AIST TSUKUBA 6 10 | Address | 31 | 92% | 3% | 12 |
| 2 | LEUCINE RESPONSIVE REGULATORY PROTEIN | Author keyword | 24 | 91% | 2% | 10 |
| 3 | ASNC | Author keyword | 21 | 90% | 2% | 9 |
| 4 | FFRP | Author keyword | 20 | 100% | 2% | 9 |
| 5 | GCVA | Author keyword | 14 | 100% | 2% | 7 |
| 6 | NIBHT | Address | 11 | 65% | 3% | 11 |
| 7 | TSUKUBA 6 10 | Address | 8 | 100% | 1% | 5 |
| 8 | DNA ADENINE METHYLASE | Author keyword | 6 | 80% | 1% | 4 |
| 9 | DO AL PROGRAM AGR SCI | Address | 6 | 100% | 1% | 4 |
| 10 | GLYCINE CLEAVAGE | Author keyword | 4 | 67% | 1% | 4 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LEUCINE RESPONSIVE REGULATORY PROTEIN | 24 | 91% | 2% | 10 | Search LEUCINE+RESPONSIVE+REGULATORY+PROTEIN | Search LEUCINE+RESPONSIVE+REGULATORY+PROTEIN |
| 2 | ASNC | 21 | 90% | 2% | 9 | Search ASNC | Search ASNC |
| 3 | FFRP | 20 | 100% | 2% | 9 | Search FFRP | Search FFRP |
| 4 | GCVA | 14 | 100% | 2% | 7 | Search GCVA | Search GCVA |
| 5 | DNA ADENINE METHYLASE | 6 | 80% | 1% | 4 | Search DNA+ADENINE+METHYLASE | Search DNA+ADENINE+METHYLASE |
| 6 | GLYCINE CLEAVAGE | 4 | 67% | 1% | 4 | Search GLYCINE+CLEAVAGE | Search GLYCINE+CLEAVAGE |
| 7 | GATC | 4 | 75% | 1% | 3 | Search GATC | Search GATC |
| 8 | HYPER THERMOPHILE | 3 | 57% | 1% | 4 | Search HYPER+THERMOPHILE | Search HYPER+THERMOPHILE |
| 9 | GCVTHP | 3 | 100% | 1% | 3 | Search GCVTHP | Search GCVTHP |
| 10 | LRP ASNC FAMILY | 3 | 100% | 1% | 3 | Search LRP+ASNC+FAMILY | Search LRP+ASNC+FAMILY |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PYELONEPHRITIS ASSOCIATED PILI | 55 | 78% | 9% | 36 |
| 2 | FFRPS | 48 | 100% | 4% | 17 |
| 3 | ESCHERICHIA COLI LRP | 41 | 87% | 5% | 20 |
| 4 | ILVIH OPERON | 40 | 82% | 5% | 23 |
| 5 | EPIGENETIC SWITCH | 23 | 100% | 2% | 10 |
| 6 | CLEAVAGE ENZYME SYSTEM | 23 | 74% | 4% | 17 |
| 7 | RESPONSIVE REGULATORY PROTEIN | 17 | 38% | 9% | 36 |
| 8 | FEAST FAMINE REGULATORY PROTEIN | 15 | 88% | 2% | 7 |
| 9 | PAP PHASE VARIATION | 15 | 73% | 3% | 11 |
| 10 | POT0434017 | 14 | 100% | 2% | 7 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| DNA methylation in bacteria: from the methyl group to the methylome | 2015 | 2 | 54 | 41% |
| The Lrp family of transcriptional regulators | 2003 | 138 | 53 | 64% |
| Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more | 2009 | 84 | 105 | 35% |
| Programmed Heterogeneity: Epigenetic Mechanisms in Bacteria | 2013 | 22 | 90 | 39% |
| Clocks and switches: bacterial gene regulation by DNA adenine methylation | 2008 | 34 | 52 | 52% |
| Dam methylation: coordinating cellular processes | 2005 | 108 | 46 | 22% |
| Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors | 2006 | 46 | 82 | 79% |
| Phase variation: how to create and coordinate population diversity | 2011 | 31 | 44 | 20% |
| Transcription regulation by feast/famine regulatory proteins, FFRPs in archaea and eubacteria | 2008 | 10 | 65 | 77% |
| Exploring bacterial epigenomics in the next-generation sequencing era: a new approach for an emerging frontier | 2014 | 5 | 55 | 18% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AIST TSUKUBA 6 10 | 31 | 92% | 2.8% | 12 |
| 2 | NIBHT | 11 | 65% | 2.6% | 11 |
| 3 | TSUKUBA 6 10 | 8 | 100% | 1.2% | 5 |
| 4 | DO AL PROGRAM AGR SCI | 6 | 100% | 0.9% | 4 |
| 5 | ERFELIJKHEIDSLEER MICROBIOL | 2 | 27% | 1.4% | 6 |
| 6 | DOCTORAL PROGRAM MED SCI | 2 | 17% | 2.4% | 10 |
| 7 | UNITE BIOCHIM LIPIDES INTERACT MACROMOL BIOL | 2 | 43% | 0.7% | 3 |
| 8 | AIST TSUKUBA 610 | 1 | 100% | 0.5% | 2 |
| 9 | AREA 12 | 1 | 50% | 0.5% | 2 |
| 10 | C T STRUCT BIOL | 1 | 50% | 0.5% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000149063 | DANISH ARCHAEA//ARCHAEAL VIRUS//FUSELLOVIRIDAE |
| 2 | 0.0000140034 | SERINE BIOSYNTHESIS//PHOSPHORYLATED PATHWAY//3 PHOSPHOGLYCERATE DEHYDROGENASE |
| 3 | 0.0000114512 | H NS//HISTONE LIKE PROTEIN//BACTERIAL CHROMATIN |
| 4 | 0.0000109941 | DNAA PROTEIN//DNAA//ORIC |
| 5 | 0.0000104258 | THREONINE DEHYDROGENASE//SERINE DEHYDRATASE//L THREONINE DEHYDROGENASE |
| 6 | 0.0000083606 | RESTRICTION MODIFICATION//RESTRICTION ENDONUCLEASE//RESTRICTION MODIFICATION SYSTEM |
| 7 | 0.0000083456 | CAULOBACTER//CTRA//CAULOBACTER CRESCENTUS |
| 8 | 0.0000078111 | TERMINATION OF REPLICATION//REPLICATION TERMINATOR PROTEIN//FORK ARREST |
| 9 | 0.0000076460 | UROPATHOGENIC ESCHERICHIA COLI//DONOR STRAND EXCHANGE//WOMENS INFECT DIS |
| 10 | 0.0000073927 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |