Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
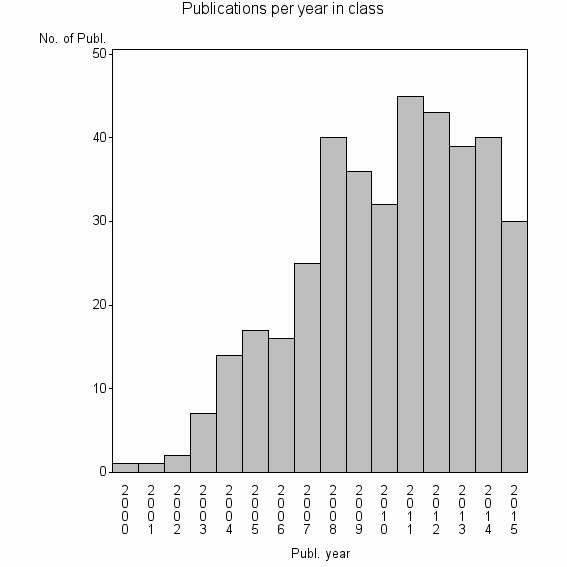
| 20708 | 388 | 46.5 | 87% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CIS MOTIF | Author keyword | 2 | 67% | 1% | 2 |
| 2 | ARABIDOPSIS T87 CELL LINE | Author keyword | 1 | 100% | 1% | 2 |
| 3 | GENE ATLAS | Author keyword | 1 | 50% | 1% | 2 |
| 4 | NAA PLANT SOIL NUTR UNIT | Address | 1 | 100% | 1% | 2 |
| 5 | GABIPD TEAM | Address | 1 | 33% | 1% | 3 |
| 6 | COEXPRESSION ANALYSIS | Author keyword | 1 | 40% | 1% | 2 |
| 7 | CO EXPRESSION ANALYSIS | Author keyword | 1 | 22% | 1% | 4 |
| 8 | GENE CO EXPRESSION NETWORKS | Author keyword | 1 | 33% | 1% | 2 |
| 9 | BIOCYC | Author keyword | 1 | 50% | 0% | 1 |
| 10 | BRAIN KOREA WORLD CLASS UNIV PROGRAM 21 | Address | 1 | 50% | 0% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CIS MOTIF | 2 | 67% | 1% | 2 | Search CIS+MOTIF | Search CIS+MOTIF |
| 2 | ARABIDOPSIS T87 CELL LINE | 1 | 100% | 1% | 2 | Search ARABIDOPSIS+T87+CELL+LINE | Search ARABIDOPSIS+T87+CELL+LINE |
| 3 | GENE ATLAS | 1 | 50% | 1% | 2 | Search GENE+ATLAS | Search GENE+ATLAS |
| 4 | COEXPRESSION ANALYSIS | 1 | 40% | 1% | 2 | Search COEXPRESSION+ANALYSIS | Search COEXPRESSION+ANALYSIS |
| 5 | CO EXPRESSION ANALYSIS | 1 | 22% | 1% | 4 | Search CO+EXPRESSION+ANALYSIS | Search CO+EXPRESSION+ANALYSIS |
| 6 | GENE CO EXPRESSION NETWORKS | 1 | 33% | 1% | 2 | Search GENE+CO+EXPRESSION+NETWORKS | Search GENE+CO+EXPRESSION+NETWORKS |
| 7 | BIOCYC | 1 | 50% | 0% | 1 | Search BIOCYC | Search BIOCYC |
| 8 | CO EXPRESSION MODULES | 1 | 50% | 0% | 1 | Search CO+EXPRESSION+MODULES | Search CO+EXPRESSION+MODULES |
| 9 | CONSERVED CO EXPRESSION | 1 | 50% | 0% | 1 | Search CONSERVED+CO+EXPRESSION | Search CONSERVED+CO+EXPRESSION |
| 10 | DROUGHT INDUCED | 1 | 50% | 0% | 1 | Search DROUGHT+INDUCED | Search DROUGHT+INDUCED |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FACTOR DATABASE | 7 | 57% | 2% | 8 |
| 2 | SATIVA SSP JAPONICA | 5 | 60% | 2% | 6 |
| 3 | MODEL DATA ANALYSIS | 5 | 26% | 5% | 18 |
| 4 | INFORMATION RESOURCE | 4 | 22% | 4% | 17 |
| 5 | ATHAMAP WEB TOOLS | 4 | 75% | 1% | 3 |
| 6 | MICROARRAY DATA SETS | 4 | 50% | 2% | 6 |
| 7 | PLANT BIOLOGY | 4 | 19% | 5% | 18 |
| 8 | ATTED II | 3 | 33% | 2% | 8 |
| 9 | COEXPRESSION NETWORKS | 3 | 21% | 3% | 13 |
| 10 | HYPOTHESIS GENERATION | 3 | 18% | 3% | 13 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Towards revealing the functions of all genes in plants | 2014 | 12 | 90 | 21% |
| Network analysis for gene discovery in plant-specialized metabolism | 2013 | 10 | 76 | 20% |
| Gene-expression analysis and network discovery using Genevestigator | 2005 | 162 | 6 | 33% |
| Co-expression and co-responses: within and beyond transcription | 2012 | 6 | 72 | 40% |
| Web Tools for Rice Transcriptome Analyses | 2011 | 4 | 19 | 47% |
| Resources for systems biology in rice | 2014 | 1 | 39 | 38% |
| Web-Queryable Large-Scale Data Sets for Hypothesis Generation in Plant Biology | 2009 | 50 | 105 | 21% |
| Approaches for extracting practical information from gene co-expression networks in plant biology | 2007 | 129 | 42 | 24% |
| Co-expression of cell wall-related genes: new tools and insights | 2012 | 9 | 55 | 33% |
| Transcriptome data modeling for targeted plant metabolic engineering | 2013 | 8 | 60 | 18% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NAA PLANT SOIL NUTR UNIT | 1 | 100% | 0.5% | 2 |
| 2 | GABIPD TEAM | 1 | 33% | 0.8% | 3 |
| 3 | BRAIN KOREA WORLD CLASS UNIV PROGRAM 21 | 1 | 50% | 0.3% | 1 |
| 4 | CROP GENOME INFORMAT | 1 | 50% | 0.3% | 1 |
| 5 | FO T OURCE CONSERVAT UTILIZAT SOUT | 1 | 50% | 0.3% | 1 |
| 6 | GENET CELLULAR DEV BIOL | 1 | 50% | 0.3% | 1 |
| 7 | GRP COMPUTAT EVOLUTIONARY BIOL | 1 | 50% | 0.3% | 1 |
| 8 | PL PLANT SCI C S | 1 | 50% | 0.3% | 1 |
| 9 | STG BASED SERV | 1 | 50% | 0.3% | 1 |
| 10 | VIRTUAL REAL PL | 1 | 50% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000193437 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 2 | 0.0000173165 | DOF TRANSCRIPTION FACTOR//DOF1//DOF PROTEIN |
| 3 | 0.0000161588 | WRKY TRANSCRIPTION FACTOR//WRKY//WRKY PROTEIN |
| 4 | 0.0000150063 | DREB//AP2 ERF//GCC BOX |
| 5 | 0.0000117831 | BIDIRECTIONAL PROMOTER//NEIGHBORING GENES//GENOME NETWORK PROJECT CORE GRPGENOM SCI |
| 6 | 0.0000117622 | METABOLITE ANNOTATION//SCRIPPS METABOL MASS SPE OMETRY//METABOLOMICS |
| 7 | 0.0000103566 | PROTEIN INTERACTION NETWORKS//PROTEIN PROTEIN INTERACTION NETWORKS//PROTEIN PROTEIN INTERACTION NETWORK |
| 8 | 0.0000101546 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 9 | 0.0000100657 | CIRCOS//BIODADOS//ORTHOLOGY |
| 10 | 0.0000096122 | JOURNAL OF BIOMEDICAL SEMANTICS//ZFIN//ZEBRAFISH INFORMAT NETWORK |