Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
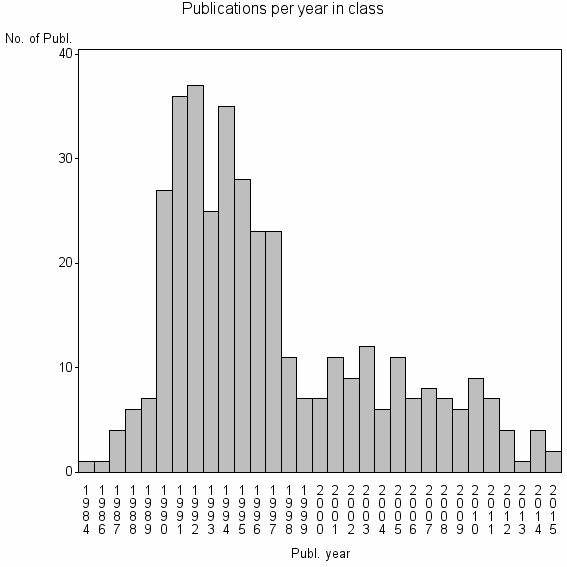
| 20862 | 382 | 21.8 | 72% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | HAPPY MAPPING | Author keyword | 3 | 100% | 1% | 3 |
| 2 | MULTILOCUS ORDERING | Author keyword | 3 | 100% | 1% | 3 |
| 3 | EXOGAMIC POPULATIONS | Author keyword | 2 | 67% | 1% | 2 |
| 4 | RADIATION HYBRID MAPS | Author keyword | 2 | 67% | 1% | 2 |
| 5 | LOCUS ORDERING | Author keyword | 2 | 50% | 1% | 3 |
| 6 | LOCUS ORDER | Author keyword | 1 | 100% | 1% | 2 |
| 7 | MARKER ORDERING | Author keyword | 1 | 100% | 1% | 2 |
| 8 | NIMH NEUROSCI | Address | 1 | 50% | 1% | 2 |
| 9 | RAPID CHAIN DELINEATION | Author keyword | 1 | 100% | 1% | 2 |
| 10 | GENE ORDERING | Author keyword | 1 | 33% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HAPPY MAPPING | 3 | 100% | 1% | 3 | Search HAPPY+MAPPING | Search HAPPY+MAPPING |
| 2 | MULTILOCUS ORDERING | 3 | 100% | 1% | 3 | Search MULTILOCUS+ORDERING | Search MULTILOCUS+ORDERING |
| 3 | EXOGAMIC POPULATIONS | 2 | 67% | 1% | 2 | Search EXOGAMIC+POPULATIONS | Search EXOGAMIC+POPULATIONS |
| 4 | RADIATION HYBRID MAPS | 2 | 67% | 1% | 2 | Search RADIATION+HYBRID+MAPS | Search RADIATION+HYBRID+MAPS |
| 5 | LOCUS ORDERING | 2 | 50% | 1% | 3 | Search LOCUS+ORDERING | Search LOCUS+ORDERING |
| 6 | LOCUS ORDER | 1 | 100% | 1% | 2 | Search LOCUS+ORDER | Search LOCUS+ORDER |
| 7 | MARKER ORDERING | 1 | 100% | 1% | 2 | Search MARKER+ORDERING | Search MARKER+ORDERING |
| 8 | RAPID CHAIN DELINEATION | 1 | 100% | 1% | 2 | Search RAPID+CHAIN+DELINEATION | Search RAPID+CHAIN+DELINEATION |
| 9 | GENE ORDERING | 1 | 33% | 1% | 2 | Search GENE+ORDERING | Search GENE+ORDERING |
| 10 | 3 TERMINAL NUCLEOTIDE | 1 | 50% | 0% | 1 | Search 3++TERMINAL+NUCLEOTIDE | Search 3++TERMINAL+NUCLEOTIDE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CONSORTIUM LINKAGE MAP | 2 | 67% | 1% | 2 |
| 2 | LOCATION DATABASE | 2 | 67% | 1% | 2 |
| 3 | MAPPING FUNCTION | 2 | 67% | 1% | 2 |
| 4 | REDUCED HYBRIDS | 2 | 67% | 1% | 2 |
| 5 | LINKED LOCI | 2 | 19% | 2% | 7 |
| 6 | NDRG4 | 1 | 50% | 1% | 2 |
| 7 | DISTAL REGION | 1 | 17% | 1% | 4 |
| 8 | 1989 ALLEN | 1 | 50% | 0% | 1 |
| 9 | IRRADIATION HYBRIDS | 1 | 50% | 0% | 1 |
| 10 | RADIATION HYBRID MAPS | 1 | 50% | 0% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Physical education - new technologies for mapping plant genomes | 2002 | 5 | 9 | 44% |
| Mapping of mammalian genomes with radiation (Goss and Harris) hybrids | 1995 | 14 | 63 | 37% |
| RADIATION HYBRIDS - IRRADIATION AND FUSION GENE-TRANSFER | 1993 | 35 | 27 | 44% |
| Linkage methods in human genetics before the computer | 2005 | 4 | 6 | 33% |
| Old can be new again: HAPPY whole genome sequencing, mapping and assembly | 2009 | 4 | 19 | 21% |
| THE END OF AN ERA | 1995 | 2 | 7 | 29% |
| An approach to high-throughput genotyping | 1996 | 35 | 23 | 9% |
| Principles, requirements and prospects of genetic mapping in plants | 2006 | 15 | 120 | 4% |
| GENETIC EPIDEMIOLOGY | 1993 | 14 | 64 | 9% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NIMH NEUROSCI | 1 | 50% | 0.5% | 2 |
| 2 | HUMAN GENET MGC | 1 | 33% | 0.5% | 2 |
| 3 | QUEENSLAND GRAINS | 1 | 50% | 0.3% | 1 |
| 4 | WAGENINGEN UR BIOMETRY | 1 | 50% | 0.3% | 1 |
| 5 | CNRS UMR GENET DEV 6061 | 0 | 33% | 0.3% | 1 |
| 6 | OXFORD MOL GRP | 0 | 33% | 0.3% | 1 |
| 7 | PROGRAM ECOSCI | 0 | 33% | 0.3% | 1 |
| 8 | UNIT GENOM | 0 | 33% | 0.3% | 1 |
| 9 | COMPUTAT INFORMAT FOOD FUTU FLAGSHI | 0 | 25% | 0.3% | 1 |
| 10 | PRINTED CIRCUIT BOARD TORY | 0 | 25% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000214042 | BIBAC//TAR CLONING//BIBAC LIBRARY |
| 2 | 0.0000159902 | DOUBLE ANEUPLOIDY//NONDISJUNCTION//48 XXY 21 |
| 3 | 0.0000149343 | MRC HUMAN ECOGENET UNIT//BIOCHEM UNIV PLEIN 1//BORN BUNGE FDN NEUROL NEUROPATHOL |
| 4 | 0.0000128657 | RELATIONSHIP ESTIMATION//PEDIGREE RECONSTRUCTION//RELATIONSHIP INFERENCE |
| 5 | 0.0000116223 | ANIM CYTOGENET GENE M PING//ANIMAL GENETICS//BIOCHEM GENET CYTOGENET |
| 6 | 0.0000091594 | GENETIC EPIDEMIOLOGY//LINKAGE ANALYSIS//MODEL FREE LINKAGE ANALYSIS |
| 7 | 0.0000078904 | RBM5//HUMAN CHROMOSOME 3//LUCA 15 |
| 8 | 0.0000074515 | FO T GENET//SYST INTEGRAT BIOL//ARBOREA CANADA CHAIR FO T ENVIRONM GENO |
| 9 | 0.0000073545 | INTERVAL MAPPING//SYSTEMS MAPPING//SELECTIVE GENOTYPING |
| 10 | 0.0000070433 | LONG GLUME//TRITICUM POLONICUM//TRITICUM PETROPAVLOVSKYI |