Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
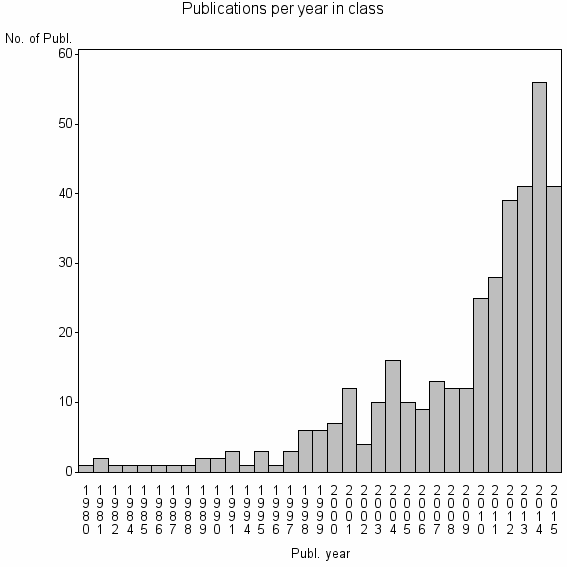
| 21159 | 371 | 44.8 | 89% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 201 | 20439 | MELAS//MITOCHONDRIAL DNA//LEBERS HEREDITARY OPTIC NEUROPATHY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | DAP3 | Author keyword | 17 | 72% | 4% | 13 |
| 2 | ICT1 | Author keyword | 6 | 80% | 1% | 4 |
| 3 | MITOCHONDRIAL RIBOSOMAL PROTEINS | Author keyword | 6 | 71% | 1% | 5 |
| 4 | MITOCHONDRIAL TRANSLATION | Author keyword | 6 | 32% | 4% | 16 |
| 5 | LRPPRC | Author keyword | 3 | 50% | 1% | 5 |
| 6 | BMRP | Author keyword | 3 | 100% | 1% | 3 |
| 7 | MITOCHONDRIAL AMINOACYL TRNA SYNTHETASE | Author keyword | 3 | 100% | 1% | 3 |
| 8 | MLASA | Author keyword | 3 | 100% | 1% | 3 |
| 9 | YARS2 | Author keyword | 3 | 100% | 1% | 3 |
| 10 | MITORIBOSOME | Author keyword | 3 | 45% | 1% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DAP3 | 17 | 72% | 4% | 13 | Search DAP3 | Search DAP3 |
| 2 | ICT1 | 6 | 80% | 1% | 4 | Search ICT1 | Search ICT1 |
| 3 | MITOCHONDRIAL RIBOSOMAL PROTEINS | 6 | 71% | 1% | 5 | Search MITOCHONDRIAL+RIBOSOMAL+PROTEINS | Search MITOCHONDRIAL+RIBOSOMAL+PROTEINS |
| 4 | MITOCHONDRIAL TRANSLATION | 6 | 32% | 4% | 16 | Search MITOCHONDRIAL+TRANSLATION | Search MITOCHONDRIAL+TRANSLATION |
| 5 | LRPPRC | 3 | 50% | 1% | 5 | Search LRPPRC | Search LRPPRC |
| 6 | BMRP | 3 | 100% | 1% | 3 | Search BMRP | Search BMRP |
| 7 | MITOCHONDRIAL AMINOACYL TRNA SYNTHETASE | 3 | 100% | 1% | 3 | Search MITOCHONDRIAL+AMINOACYL+TRNA+SYNTHETASE | Search MITOCHONDRIAL+AMINOACYL+TRNA+SYNTHETASE |
| 8 | MLASA | 3 | 100% | 1% | 3 | Search MLASA | Search MLASA |
| 9 | YARS2 | 3 | 100% | 1% | 3 | Search YARS2 | Search YARS2 |
| 10 | MITORIBOSOME | 3 | 45% | 1% | 5 | Search MITORIBOSOME | Search MITORIBOSOME |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DAP3 | 34 | 93% | 4% | 13 |
| 2 | CORRESPONDING GENE SEQUENCES | 24 | 82% | 4% | 14 |
| 3 | DEATH ASSOCIATED PROTEIN 3 | 19 | 76% | 4% | 13 |
| 4 | SIDEROBLASTIC ANEMIA MLASA | 14 | 50% | 5% | 20 |
| 5 | PERRAULT SYNDROME | 7 | 38% | 4% | 14 |
| 6 | HSUV3P HELICASE | 6 | 80% | 1% | 4 |
| 7 | LRP130 | 6 | 80% | 1% | 4 |
| 8 | LACTATE ELEVATION | 4 | 67% | 1% | 4 |
| 9 | LRPPRC | 4 | 67% | 1% | 4 |
| 10 | ELONGATION FACTOR EFTS | 4 | 75% | 1% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Mitochondrial protein synthesis: Figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation | 2014 | 9 | 108 | 50% |
| Human diseases with impaired mitochondrial protein synthesis | 2011 | 50 | 57 | 46% |
| Making Proteins in the Powerhouse | 2014 | 15 | 139 | 29% |
| Mitochondrial aminoacyl-tRNA synthetases in human disease | 2013 | 22 | 34 | 29% |
| Properties of human mitochondrial ribosomes | 2003 | 66 | 29 | 76% |
| Supernumerary proteins of mitochondrial ribosomes | 2014 | 2 | 60 | 57% |
| Human pentatricopeptide proteins Only a few and what do they do? | 2013 | 2 | 66 | 42% |
| The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression | 2012 | 10 | 165 | 32% |
| The role of mammalian PPR domain proteins in the regulation of mitochondrial gene expression | 2012 | 7 | 86 | 35% |
| The 55S mammalian mitochondrial ribosome and its tRNA-exit region | 2015 | 0 | 52 | 40% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | WELLCOME TRUST MITOCHONDRIAL | 2 | 13% | 4.3% | 16 |
| 2 | AG K S | 2 | 67% | 0.5% | 2 |
| 3 | CARDIFF UNIV PEKING UNIV ONCOL JOINT | 2 | 67% | 0.5% | 2 |
| 4 | CUPUCI | 1 | 50% | 0.5% | 2 |
| 5 | PROGRAMS UNIT MOL NEUROL | 1 | 50% | 0.5% | 2 |
| 6 | GENET METAB DISORDERS UNIT | 1 | 25% | 1.1% | 4 |
| 7 | PUBL HLTH ENVIRONM SCI ENGN | 1 | 23% | 0.8% | 3 |
| 8 | ANDROGEN RECEPTOR | 1 | 50% | 0.3% | 1 |
| 9 | CHUPS | 1 | 50% | 0.3% | 1 |
| 10 | COMPUTAT MOL BIOL PROGRAMME | 1 | 50% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000288256 | MITOCHONDRIAL NUCLEOIDS//TFAM//MITOCHONDRIAL NUCLEOID |
| 2 | 0.0000249665 | YIDC//OXA1//COX17 |
| 3 | 0.0000124511 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 4 | 0.0000121651 | PODOSPORA ANSERINA//LINEAR MITOCHONDRIAL PLASMID//KALILO |
| 5 | 0.0000114746 | BLUE NATIVE ELECTROPHORESIS//TMEM70//BCS1L |
| 6 | 0.0000112958 | MITO PORTER//MITOCHONDRIAL DRUG DELIVERY//MITOCHONDRIAL GENE THERAPY |
| 7 | 0.0000107001 | OBG//DRG2//BACTERIAL GTPASE |
| 8 | 0.0000098938 | COMPLEX I//NADH UBIQUINONE OXIDOREDUCTASE//NADH UBIQUINONE OXIDOREDUCTASE |
| 9 | 0.0000097483 | TRNA NUCLEOTIDYLTRANSFERASE//CCA ADDING ENZYME//TRNAHIS GUANYLYLTRANSFERASE |
| 10 | 0.0000094353 | POLG//MNGIE//DEOXYRIBONUCLEOSIDE KINASE |