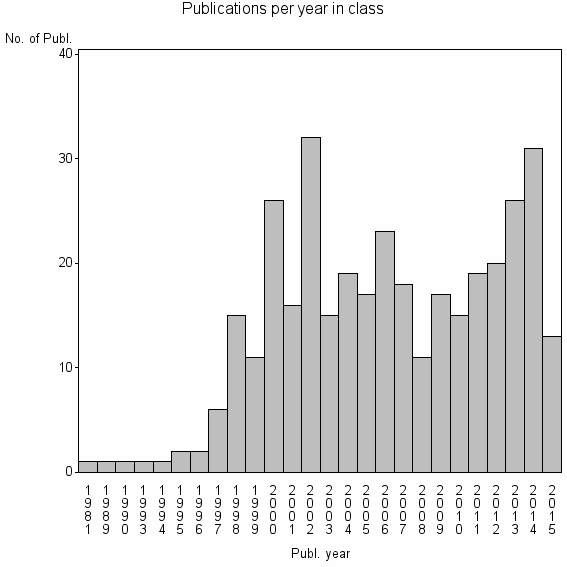
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 21496 | 359 | 43.1 | 83% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2844 | 2448 | PEPTIDE DEFORMYLASE//ENOYL ACP REDUCTASE//PLATENSIMYCIN |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | ESSENTIAL GENES | Author keyword | 4 | 20% | 6% | 20 |
| 2 | SUBTRACTIVE GENOMICS | Author keyword | 4 | 56% | 1% | 5 |
| 3 | ESSENTIAL GENE | Author keyword | 2 | 18% | 3% | 12 |
| 4 | ANTIMICROBIALS HOST DEF CEDD | Address | 2 | 67% | 1% | 2 |
| 5 | ALLELIC REPLACEMENT MUTAGENESIS | Author keyword | 1 | 100% | 1% | 2 |
| 6 | COMPUTATIONAL COMPARATIVE GENOMICS | Author keyword | 1 | 100% | 1% | 2 |
| 7 | ESSENTIAL GENE PREDICTION | Author keyword | 1 | 100% | 1% | 2 |
| 8 | IN SILICO DRUG TARGETS | Author keyword | 1 | 100% | 1% | 2 |
| 9 | KAAS SERVER | Author keyword | 1 | 100% | 1% | 2 |
| 10 | NEW ANTIMICROBIAL AGENT | Author keyword | 1 | 100% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ESSENTIAL GENES | 4 | 20% | 6% | 20 | Search ESSENTIAL+GENES | Search ESSENTIAL+GENES |
| 2 | SUBTRACTIVE GENOMICS | 4 | 56% | 1% | 5 | Search SUBTRACTIVE+GENOMICS | Search SUBTRACTIVE+GENOMICS |
| 3 | ESSENTIAL GENE | 2 | 18% | 3% | 12 | Search ESSENTIAL+GENE | Search ESSENTIAL+GENE |
| 4 | ALLELIC REPLACEMENT MUTAGENESIS | 1 | 100% | 1% | 2 | Search ALLELIC+REPLACEMENT+MUTAGENESIS | Search ALLELIC+REPLACEMENT+MUTAGENESIS |
| 5 | COMPUTATIONAL COMPARATIVE GENOMICS | 1 | 100% | 1% | 2 | Search COMPUTATIONAL+COMPARATIVE+GENOMICS | Search COMPUTATIONAL+COMPARATIVE+GENOMICS |
| 6 | ESSENTIAL GENE PREDICTION | 1 | 100% | 1% | 2 | Search ESSENTIAL+GENE+PREDICTION | Search ESSENTIAL+GENE+PREDICTION |
| 7 | IN SILICO DRUG TARGETS | 1 | 100% | 1% | 2 | Search IN+SILICO+DRUG+TARGETS | Search IN+SILICO+DRUG+TARGETS |
| 8 | KAAS SERVER | 1 | 100% | 1% | 2 | Search KAAS+SERVER | Search KAAS+SERVER |
| 9 | NEW ANTIMICROBIAL AGENT | 1 | 100% | 1% | 2 | Search NEW+ANTIMICROBIAL+AGENT | Search NEW+ANTIMICROBIAL+AGENT |
| 10 | XYL TET | 1 | 100% | 1% | 2 | Search XYL+TET | Search XYL+TET |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ESSENTIAL BACTERIAL GENES | 21 | 69% | 5% | 18 |
| 2 | GLOBAL TRANSPOSON MUTAGENESIS | 17 | 68% | 4% | 15 |
| 3 | ALLELIC REPLACEMENT MUTAGENESIS | 12 | 86% | 2% | 6 |
| 4 | ESSENTIAL GENES | 9 | 19% | 12% | 42 |
| 5 | DEG | 3 | 50% | 1% | 5 |
| 6 | TRANSPOSON MUTANT LIBRARY | 3 | 31% | 3% | 9 |
| 7 | VACCINE TARGETS | 2 | 38% | 1% | 5 |
| 8 | CONDITIONALLY ESSENTIAL GENES | 2 | 32% | 2% | 6 |
| 9 | ANTIMICROBIAL TARGETS | 2 | 67% | 1% | 2 |
| 10 | PROTEIN REDUCTASE FABI | 2 | 67% | 1% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Essential genes on metabolic maps | 2006 | 58 | 48 | 42% |
| Transposon-based approaches to identify essential bacterial genes | 2000 | 48 | 13 | 69% |
| Essence of life: essential genes of minimal genomes | 2011 | 37 | 54 | 24% |
| The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antibacterials | 2007 | 40 | 80 | 30% |
| How many genes can make a cell: The minimal-gene-set concept | 2000 | 128 | 24 | 25% |
| From essential to persistent genes: a functional approach to constructing synthetic life | 2013 | 7 | 64 | 31% |
| Determination of the core of a minimal bacterial gene set | 2004 | 237 | 80 | 14% |
| Building the blueprint of life | 2010 | 8 | 46 | 35% |
| Bacterial genomics and pathogen evolution | 2006 | 68 | 68 | 16% |
| Essential genes as antimicrobial targets and cornerstones of synthetic biology | 2012 | 10 | 68 | 25% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ANTIMICROBIALS HOST DEF CEDD | 2 | 67% | 0.6% | 2 |
| 2 | PATHOGEN GENET | 1 | 50% | 0.6% | 2 |
| 3 | ANTIBACTERIAL | 1 | 33% | 0.6% | 2 |
| 4 | AFFILIATED STAFF BIOINFORMAT | 1 | 50% | 0.3% | 1 |
| 5 | BIOMED ELECT BIOINFORMAT SYST BIO | 1 | 50% | 0.3% | 1 |
| 6 | JAMIL UR RAHMAN GENOME | 1 | 50% | 0.3% | 1 |
| 7 | CHEMOTHER Y MOL GENET | 1 | 25% | 0.6% | 2 |
| 8 | 46Y | 0 | 33% | 0.3% | 1 |
| 9 | BUR AGR IMPORTANT MICROORGANISMS | 0 | 33% | 0.3% | 1 |
| 10 | GENET VEGETALE MOULON | 0 | 33% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000135135 | SYNTHETIC DOSAGE LETHALITY//YEAST DELETION//SYNTHETIC GENETIC ARRAY |
| 2 | 0.0000130566 | RECOMBINEERING//LAMBDA RED//LAMBDA RED RECOMBINATION |
| 3 | 0.0000117830 | PLATENSIMYCIN//CORTISTATIN A//PLATENCIN |
| 4 | 0.0000113693 | OBG//DRG2//BACTERIAL GTPASE |
| 5 | 0.0000109833 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 6 | 0.0000101606 | PEPTIDE DEFORMYLASE//PEPTIDE DEFORMYLASE INHIBITOR//DEFORMYLASE |
| 7 | 0.0000096040 | ALANINE RACEMASE//MURD//GLUTAMATE RACEMASE |
| 8 | 0.0000087156 | FLUX BALANCE ANALYSIS//CONSTRAINT BASED MODELING//METABOLIC NETWORK |
| 9 | 0.0000084891 | SYNTHETIC BIOLOGY//ACS SYNTHETIC BIOLOGY//CALIF QUANTITAT BIOL QB3 |
| 10 | 0.0000074895 | GENETIC INSTRUCTIONS//THERMOSYNTHESIS//EXCLUSION ZONE EZ |