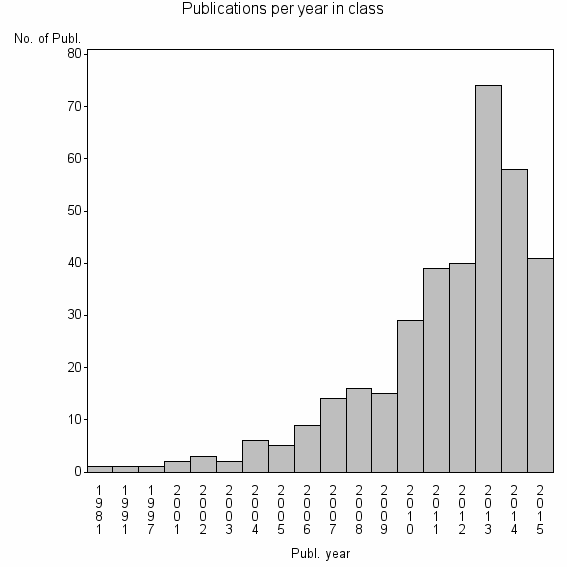
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 21583 | 356 | 39.0 | 87% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1568 | 6715 | NEXT GENERATION SEQUENCING//RNA SEQ//COPY NUMBER VARIATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EPIDEMIOL PATHOPHYSIOL ONCOGEN VIRUS UNIT | Address | 3 | 100% | 1% | 3 |
| 2 | LYSSAVIRUS DYNAM HOST AD TAT UNIT | Address | 3 | 50% | 1% | 4 |
| 3 | VIRAL DISCOVERY | Author keyword | 3 | 60% | 1% | 3 |
| 4 | VIRUS DISCOVERY | Author keyword | 2 | 20% | 3% | 10 |
| 5 | SEQUENCING ERROR CORRECTION | Author keyword | 2 | 67% | 1% | 2 |
| 6 | VIRAL METAGENOMICS | Author keyword | 2 | 20% | 2% | 7 |
| 7 | CLIN VIROL MED MICROBIOL | Address | 1 | 50% | 1% | 2 |
| 8 | MASSTAG PCR | Author keyword | 1 | 50% | 1% | 2 |
| 9 | VIRAL EMERGENT DIS UNIT | Address | 1 | 100% | 1% | 2 |
| 10 | RESEQUENCING MICROARRAY | Author keyword | 1 | 33% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | VIRAL DISCOVERY | 3 | 60% | 1% | 3 | Search VIRAL+DISCOVERY | Search VIRAL+DISCOVERY |
| 2 | VIRUS DISCOVERY | 2 | 20% | 3% | 10 | Search VIRUS+DISCOVERY | Search VIRUS+DISCOVERY |
| 3 | SEQUENCING ERROR CORRECTION | 2 | 67% | 1% | 2 | Search SEQUENCING+ERROR+CORRECTION | Search SEQUENCING+ERROR+CORRECTION |
| 4 | VIRAL METAGENOMICS | 2 | 20% | 2% | 7 | Search VIRAL+METAGENOMICS | Search VIRAL+METAGENOMICS |
| 5 | MASSTAG PCR | 1 | 50% | 1% | 2 | Search MASSTAG+PCR | Search MASSTAG+PCR |
| 6 | RESEQUENCING MICROARRAY | 1 | 33% | 1% | 3 | Search RESEQUENCING+MICROARRAY | Search RESEQUENCING+MICROARRAY |
| 7 | PATHOGEN DISCOVERY | 1 | 25% | 1% | 4 | Search PATHOGEN+DISCOVERY | Search PATHOGEN+DISCOVERY |
| 8 | VIDISCA | 1 | 33% | 1% | 2 | Search VIDISCA | Search VIDISCA |
| 9 | ADVENTITIOUS AGENT | 1 | 50% | 0% | 1 | Search ADVENTITIOUS+AGENT | Search ADVENTITIOUS+AGENT |
| 10 | BASE CALL ERROR | 1 | 50% | 0% | 1 | Search BASE+CALL+ERROR | Search BASE+CALL+ERROR |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | VIRUS DISCOVERY | 11 | 78% | 2% | 7 |
| 2 | QUASI SPECIES RECONSTRUCTION | 6 | 80% | 1% | 4 |
| 3 | SINGLE PRIMER AMPLIFICATION | 4 | 75% | 1% | 3 |
| 4 | VIRUS IDENTIFICATION | 3 | 60% | 1% | 3 |
| 5 | GENERATION SEQUENCING DATA | 3 | 16% | 4% | 16 |
| 6 | RESEQUENCING MICROARRAY | 2 | 67% | 1% | 2 |
| 7 | VIRAL METAGENOMICS | 2 | 23% | 2% | 8 |
| 8 | MICROBIAL DETECTION | 1 | 50% | 1% | 2 |
| 9 | OIE COLLABORATING CENTER | 1 | 100% | 1% | 2 |
| 10 | VIRAL PATHOGENS | 1 | 14% | 3% | 9 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Next-generation sequencing technology in clinical virology | 2013 | 34 | 67 | 27% |
| Viral pathogen discovery | 2013 | 24 | 96 | 26% |
| Metagenomic identification of viral pathogens | 2013 | 14 | 43 | 33% |
| Application of next-generation sequencing technologies in virology | 2012 | 56 | 92 | 16% |
| Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data | 2012 | 43 | 156 | 19% |
| Application of resequencing microarrays in microbial detection and characterization | 2012 | 4 | 77 | 44% |
| Applications of Next-Generation Sequencing Technologies to Diagnostic Virology | 2011 | 51 | 130 | 18% |
| Metagenomics for the discovery of novel human viruses | 2010 | 55 | 130 | 15% |
| Next-generation sequencing technologies in diagnostic virology | 2013 | 14 | 25 | 20% |
| Metagenomic analysis of bacterial infections by means of high-throughput DNA sequencing | 2011 | 13 | 12 | 25% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EPIDEMIOL PATHOPHYSIOL ONCOGEN VIRUS UNIT | 3 | 100% | 0.8% | 3 |
| 2 | LYSSAVIRUS DYNAM HOST AD TAT UNIT | 3 | 50% | 1.1% | 4 |
| 3 | CLIN VIROL MED MICROBIOL | 1 | 50% | 0.6% | 2 |
| 4 | VIRAL EMERGENT DIS UNIT | 1 | 100% | 0.6% | 2 |
| 5 | URGENT PONSES BIOL THREATS | 1 | 33% | 0.8% | 3 |
| 6 | CNRS URA3015 | 1 | 40% | 0.6% | 2 |
| 7 | BACTERIAL MOL GENET UNIT | 1 | 22% | 1.1% | 4 |
| 8 | ABORATING BIOTECHNOL BASED DIAG INFECT DI | 1 | 33% | 0.6% | 2 |
| 9 | URGENT PONSE BIOL THREATS CIBU | 1 | 33% | 0.6% | 2 |
| 10 | ANIM BREEDING GENET HGEN | 1 | 50% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000179851 | METAGENOMICS//MICROBIAL GENOM METAGEN PROGRAM//BIOSCI RD |
| 2 | 0.0000167807 | GENOME ASSEMBLY//VARIANT CALLING//TARGET ENRICHMENT |
| 3 | 0.0000155996 | NONSPECIFIC HYBRIDIZATIONS//NEAREST NEIGHBOR PROPERTIES//NUCLEIC ACID OLIGOMERS |
| 4 | 0.0000128468 | HUMAN BOCAVIRUS//BOCAVIRUS//HBOV |
| 5 | 0.0000113823 | ASTROVIRUS//PICOBIRNAVIRUS//TURKEY ASTROVIRUS |
| 6 | 0.0000112252 | RAPID INFLUENZA DIAGNOSTIC TEST//RESPIRATORY VIRUSES//RAPID ANTIGEN TEST |
| 7 | 0.0000101608 | CULEX FLAVIVIRUS//TEMBUSU VIRUS//DUCK TEMBUSU VIRUS |
| 8 | 0.0000086691 | XMRV//XENOTROPIC MURINE LEUKEMIA VIRUS RELATED VIRUS//XENOTROPIC MURINE LEUKEMIA VIRUS RELATED VIRUS XMRV |
| 9 | 0.0000078172 | K65R//TRANSMITTED DRUG RESISTANCE//HIV 1 DRUG RESISTANCE |
| 10 | 0.0000069532 | WHOLE GENOME AMPLIFICATION//MULTIPLE DISPLACEMENT AMPLIFICATION//PHI29 DNA POLYMERASE |