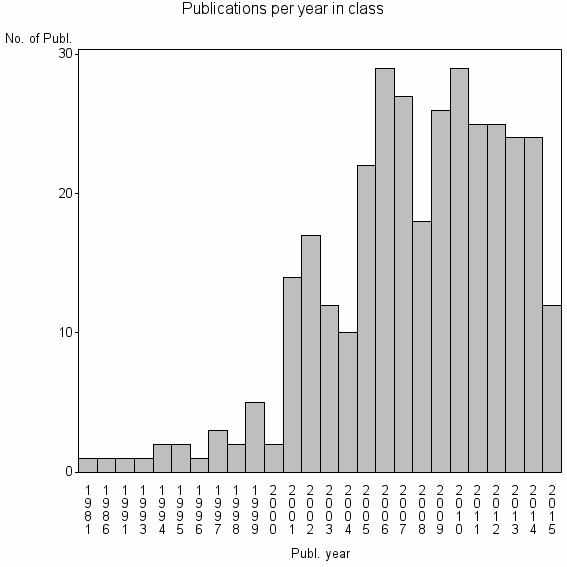
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22133 | 335 | 42.3 | 80% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EVOLUTIONARY BIOINFORMAT | Address | 18 | 59% | 6% | 20 |
| 2 | CLOSED LOOPS | Author keyword | 13 | 50% | 6% | 19 |
| 3 | SKEW PAIRS | Author keyword | 6 | 100% | 1% | 4 |
| 4 | GENOME ERS | Address | 5 | 17% | 8% | 28 |
| 5 | EARLIEST PROTEINS | Author keyword | 4 | 75% | 1% | 3 |
| 6 | FOLD SUPERFAMILY | Author keyword | 4 | 75% | 1% | 3 |
| 7 | CONSERVED SEQUENCE STRUCTURE MODULES | Author keyword | 3 | 100% | 1% | 3 |
| 8 | EVOLUTION OF DOMAIN ARCHITECTURE | Author keyword | 3 | 100% | 1% | 3 |
| 9 | MAJOR FOLDS | Author keyword | 3 | 100% | 1% | 3 |
| 10 | MODULAR PROTEIN EVOLUTION | Author keyword | 3 | 100% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CLOSED LOOPS | 13 | 50% | 6% | 19 | Search CLOSED+LOOPS | Search CLOSED+LOOPS |
| 2 | SKEW PAIRS | 6 | 100% | 1% | 4 | Search SKEW+PAIRS | Search SKEW+PAIRS |
| 3 | EARLIEST PROTEINS | 4 | 75% | 1% | 3 | Search EARLIEST+PROTEINS | Search EARLIEST+PROTEINS |
| 4 | FOLD SUPERFAMILY | 4 | 75% | 1% | 3 | Search FOLD+SUPERFAMILY | Search FOLD+SUPERFAMILY |
| 5 | CONSERVED SEQUENCE STRUCTURE MODULES | 3 | 100% | 1% | 3 | Search CONSERVED+SEQUENCE+STRUCTURE+MODULES | Search CONSERVED+SEQUENCE+STRUCTURE+MODULES |
| 6 | EVOLUTION OF DOMAIN ARCHITECTURE | 3 | 100% | 1% | 3 | Search EVOLUTION+OF+DOMAIN+ARCHITECTURE | Search EVOLUTION+OF+DOMAIN+ARCHITECTURE |
| 7 | MAJOR FOLDS | 3 | 100% | 1% | 3 | Search MAJOR+FOLDS | Search MAJOR+FOLDS |
| 8 | MODULAR PROTEIN EVOLUTION | 3 | 100% | 1% | 3 | Search MODULAR+PROTEIN+EVOLUTION | Search MODULAR+PROTEIN+EVOLUTION |
| 9 | ORGANISMAL DIVERSIFICATION | 3 | 60% | 1% | 3 | Search ORGANISMAL+DIVERSIFICATION | Search ORGANISMAL+DIVERSIFICATION |
| 10 | RIBONUCLEOPROTEIN WORLD | 3 | 60% | 1% | 3 | Search RIBONUCLEOPROTEIN+WORLD | Search RIBONUCLEOPROTEIN+WORLD |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MULTIDOMAIN PROTEINS | 12 | 27% | 11% | 38 |
| 2 | FOLD ARCHITECTURE | 12 | 86% | 2% | 6 |
| 3 | PROTEIN FOLD ARCHITECTURE | 11 | 100% | 2% | 6 |
| 4 | DOMAIN ARCHITECTURES | 3 | 57% | 1% | 4 |
| 5 | PROTEOMES | 3 | 14% | 6% | 19 |
| 6 | PHYLOGENOMIC ANALYSIS | 3 | 29% | 2% | 8 |
| 7 | CLOSED LOOPS | 3 | 32% | 2% | 7 |
| 8 | DOMAIN REARRANGEMENTS | 2 | 67% | 1% | 2 |
| 9 | NOTCH GENES | 2 | 67% | 1% | 2 |
| 10 | PROTEIN SEQUENCE SPACE | 2 | 67% | 1% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Arrangements in the modular evolution of proteins | 2008 | 70 | 64 | 33% |
| Characters of very ancient proteins | 2008 | 21 | 19 | 58% |
| Reassessing Domain Architecture Evolution of Metazoan Proteins: Major Impact of Gene Prediction Errors | 2011 | 7 | 51 | 35% |
| How do new proteins arise? | 2010 | 20 | 64 | 23% |
| Iron in evolution | 2012 | 8 | 7 | 43% |
| The evolution of domain arrangements in proteins and interaction networks | 2005 | 58 | 59 | 19% |
| Genomic and structural aspects of protein evolution | 2009 | 47 | 64 | 22% |
| Evolutionary aspects of protein structure and folding | 2003 | 36 | 45 | 29% |
| Evolution of protein modularity | 2009 | 17 | 44 | 30% |
| The Role of Domain Shuffling in the Evolution of Signaling Networks | 2014 | 0 | 34 | 29% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EVOLUTIONARY BIOINFORMAT | 18 | 59% | 6.0% | 20 |
| 2 | GENOME ERS | 5 | 17% | 8.4% | 28 |
| 3 | EVOLUTIONARY BIOINFORMAT GRP | 2 | 28% | 1.5% | 5 |
| 4 | PROGRAM COMPUTAT ORG SOC | 1 | 100% | 0.6% | 2 |
| 5 | CNRS URA 1354 GENET MICROBIOL | 1 | 50% | 0.3% | 1 |
| 6 | GENOME ERSITY | 1 | 50% | 0.3% | 1 |
| 7 | LMCP MINERAL CRISTALLOG | 1 | 50% | 0.3% | 1 |
| 8 | MATH CHEM GRP | 1 | 16% | 0.9% | 3 |
| 9 | MATH INITIAT | 0 | 33% | 0.3% | 1 |
| 10 | SCI DATA MANAGEMENT | 0 | 33% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000185360 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//CASP |
| 2 | 0.0000157358 | CIRCOS//BIODADOS//ORTHOLOGY |
| 3 | 0.0000155995 | TREE OF LIFE//HORIZONTAL GENE TRANSFER//LATERAL GENE TRANSFER |
| 4 | 0.0000132695 | INTRON GAIN//INTRON LOSS//INTRON EVOLUTION |
| 5 | 0.0000122981 | COMPLEX SCI SOFTWARE//3D ZERNIKE DESCRIPTOR//PROTEIN SURFACE SHAPE |
| 6 | 0.0000117058 | GENETIC CODE//EQUIPE BIOINFORMAT THEOR//CIRCULAR CODE |
| 7 | 0.0000115749 | EMBL OUTSTN//PROT INFORMAT OURCE//HLTH SCI TECHNOL ICIR |
| 8 | 0.0000075445 | SEX BIASED GENE EXPRESSION//SEXUAL ANTAGONISM//INTRALOCUS SEXUAL CONFLICT |
| 9 | 0.0000074187 | PROTEIN INTERACTION NETWORKS//PROTEIN PROTEIN INTERACTION NETWORKS//PROTEIN PROTEIN INTERACTION NETWORK |
| 10 | 0.0000067524 | WHOLE GENOME DUPLICATION//GENET MOL CELLULAIRE ECT LEVU INTERE//GENOME DUPLICATION |