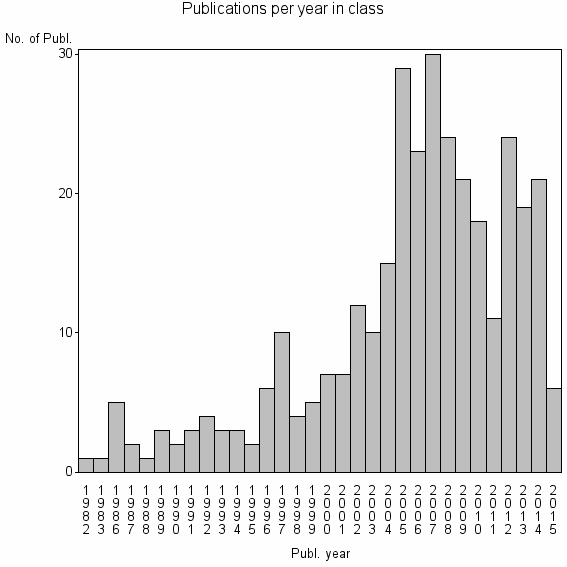
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22231 | 332 | 42.9 | 89% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | WRAP53 | Author keyword | 7 | 64% | 2% | 7 |
| 2 | OVERLAPPING GENES | Author keyword | 6 | 27% | 5% | 18 |
| 3 | CUSTOM BIOTECHNOL SERV GRP | Address | 6 | 100% | 1% | 4 |
| 4 | NATURAL ANTISENSE TRANSCRIPTS NATS | Author keyword | 4 | 75% | 1% | 3 |
| 5 | NUDT6 | Author keyword | 4 | 75% | 1% | 3 |
| 6 | NATURAL ANTISENSE TRANSCRIPT | Author keyword | 4 | 33% | 3% | 9 |
| 7 | ENDOGENOUS ANTISENSE RNA | Author keyword | 3 | 100% | 1% | 3 |
| 8 | NATURAL ANTISENSE TRANSCRIPTS | Author keyword | 3 | 33% | 2% | 8 |
| 9 | GFG | Author keyword | 2 | 67% | 1% | 2 |
| 10 | NATURAL ANTISENSE TRANSCRIPT NAT | Author keyword | 2 | 67% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | WRAP53 | 7 | 64% | 2% | 7 | Search WRAP53 | Search WRAP53 |
| 2 | OVERLAPPING GENES | 6 | 27% | 5% | 18 | Search OVERLAPPING+GENES | Search OVERLAPPING+GENES |
| 3 | NATURAL ANTISENSE TRANSCRIPTS NATS | 4 | 75% | 1% | 3 | Search NATURAL+ANTISENSE+TRANSCRIPTS+NATS | Search NATURAL+ANTISENSE+TRANSCRIPTS+NATS |
| 4 | NUDT6 | 4 | 75% | 1% | 3 | Search NUDT6 | Search NUDT6 |
| 5 | NATURAL ANTISENSE TRANSCRIPT | 4 | 33% | 3% | 9 | Search NATURAL+ANTISENSE+TRANSCRIPT | Search NATURAL+ANTISENSE+TRANSCRIPT |
| 6 | ENDOGENOUS ANTISENSE RNA | 3 | 100% | 1% | 3 | Search ENDOGENOUS+ANTISENSE+RNA | Search ENDOGENOUS+ANTISENSE+RNA |
| 7 | NATURAL ANTISENSE TRANSCRIPTS | 3 | 33% | 2% | 8 | Search NATURAL+ANTISENSE+TRANSCRIPTS | Search NATURAL+ANTISENSE+TRANSCRIPTS |
| 8 | GFG | 2 | 67% | 1% | 2 | Search GFG | Search GFG |
| 9 | NATURAL ANTISENSE TRANSCRIPT NAT | 2 | 67% | 1% | 2 | Search NATURAL+ANTISENSE+TRANSCRIPT+NAT | Search NATURAL+ANTISENSE+TRANSCRIPT+NAT |
| 10 | ANTISENSE TRANSCRIPTOME | 1 | 100% | 1% | 2 | Search ANTISENSE+TRANSCRIPTOME | Search ANTISENSE+TRANSCRIPTOME |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GFG | 6 | 80% | 1% | 4 |
| 2 | BIOINFORMATIC ANALYSIS SUGGESTS | 6 | 100% | 1% | 4 |
| 3 | OVERLAPPING GENES | 5 | 19% | 6% | 21 |
| 4 | OVERLAPPING TRANSCRIPTION | 3 | 50% | 1% | 4 |
| 5 | CONVERGENT PROMOTERS | 2 | 67% | 1% | 2 |
| 6 | PROKARYOTIC GENES | 2 | 67% | 1% | 2 |
| 7 | WRAP53 | 2 | 67% | 1% | 2 |
| 8 | RNA TRANSCRIPT | 2 | 50% | 1% | 3 |
| 9 | OPPOSITE STRAND | 2 | 15% | 3% | 11 |
| 10 | OVERLAPPING READING FRAMES | 2 | 21% | 2% | 7 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Regulatory roles of natural antisense transcripts | 2009 | 221 | 60 | 27% |
| In search of antisense | 2004 | 196 | 38 | 39% |
| Natural antisense transcripts regulate gene expression in an epigenetic manner | 2010 | 23 | 56 | 32% |
| Mechanisms of Small RNA Generation from Cis-NATs in Response to Environmental and Developmental Cues | 2013 | 8 | 77 | 26% |
| In situ Hybridization | 2012 | 0 | 6 | 67% |
| Overlapping genes in vertebrate genomes | 2005 | 51 | 88 | 38% |
| Naturally occurring antisense RNA: function and mechanisms of action | 2009 | 29 | 45 | 31% |
| Antisense transcription: A critical look in both directions | 2009 | 54 | 216 | 21% |
| Natural Antisense Transcripts in Plants: A Review and Identification in Soybean Infected with Phakopsora pachyrhizi SuperSAGE Library | 2013 | 0 | 57 | 46% |
| Overlapping transcripts, double-stranded RNA and antisense regulation: A genomic perspective | 2006 | 46 | 194 | 18% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CUSTOM BIOTECHNOL SERV GRP | 6 | 100% | 1.2% | 4 |
| 2 | TECHNOL DEV TEAM MAMMALIAN CELLULAR DYNAM | 2 | 22% | 2.4% | 8 |
| 3 | 1560 EST SHERBROOKE | 1 | 50% | 0.3% | 1 |
| 4 | ABT ANALYT BIOGEOCHEM | 1 | 50% | 0.3% | 1 |
| 5 | AG MOL INTERACT | 1 | 50% | 0.3% | 1 |
| 6 | ELE ONPHYS | 1 | 50% | 0.3% | 1 |
| 7 | PROTE SIGNAL TRASDUCT | 1 | 50% | 0.3% | 1 |
| 8 | SCI BIOL SUPRAMOL SYSTTSURUMI KU | 1 | 50% | 0.3% | 1 |
| 9 | SKLG | 1 | 50% | 0.3% | 1 |
| 10 | URA CNRS3012 | 1 | 50% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000214612 | LONG NONCODING RNAS//LNCRNA//LONG NON CODING RNA |
| 2 | 0.0000197776 | BIDIRECTIONAL PROMOTER//NEIGHBORING GENES//GENOME NETWORK PROJECT CORE GRPGENOM SCI |
| 3 | 0.0000192525 | SMALL ACTIVATING RNA//SARNA//RNA ACTIVATION |
| 4 | 0.0000157459 | PROCESSED PSEUDOGENES//PHYSICAL AND PSYCHOLOGICAL STRESSES//DUPLICATED PSEUDOGENES |
| 5 | 0.0000111719 | SERIAL ANALYSIS OF GENE EXPRESSION//SAGE//GLGI |
| 6 | 0.0000109179 | ADAR//RNA EDITING//ADAR2 |
| 7 | 0.0000081131 | GENE PREDICTION//GENE FINDING//GENE RECOGNITION |
| 8 | 0.0000054175 | SEX BIASED GENE EXPRESSION//SEXUAL ANTAGONISM//INTRALOCUS SEXUAL CONFLICT |
| 9 | 0.0000053848 | TNFAIP1//KCTD7//KCTD10 |
| 10 | 0.0000052043 | BACTERIOPHAGE ALPHA 3//MICROVIRUS//BACTERIOPHAGE PHI X174 |