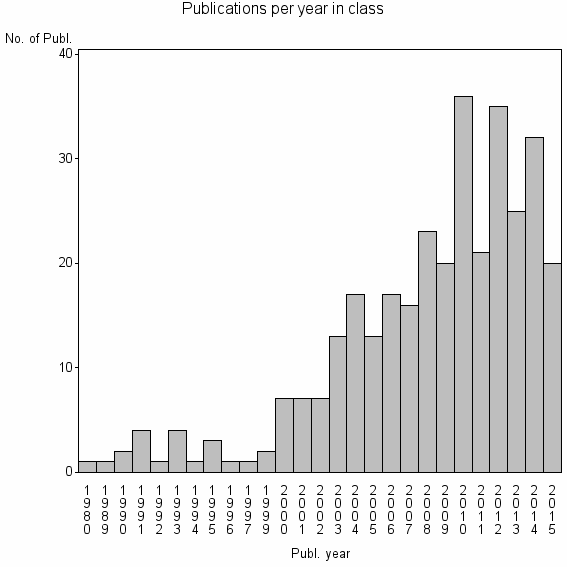
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22291 | 330 | 39.4 | 86% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1307 | 8075 | CLICK CHEMISTRY//1 2 3 TRIAZOLES//1 2 3 TRIAZOLE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PHARMACEUT CHEM BIOANALYT | Address | 14 | 48% | 6% | 21 |
| 2 | CROSS LINKING MASS SPECTROMETRY | Author keyword | 6 | 80% | 1% | 4 |
| 3 | CHEMICAL CROSS LINKING | Author keyword | 4 | 14% | 8% | 27 |
| 4 | CROSS LINKED PEPTIDES | Author keyword | 4 | 75% | 1% | 3 |
| 5 | IN VIVO PROTEIN INTERACTIONS | Author keyword | 2 | 67% | 1% | 2 |
| 6 | PROTEIN 3D STRUCTURE | Author keyword | 2 | 25% | 2% | 6 |
| 7 | PROTEIN INTERACTION REPORTER | Author keyword | 1 | 100% | 1% | 2 |
| 8 | XL MS | Author keyword | 1 | 100% | 1% | 2 |
| 9 | GENOME BRITISH COLUMBIA PROTE | Address | 1 | 16% | 2% | 8 |
| 10 | GCAP 2 | Author keyword | 1 | 33% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CROSS LINKING MASS SPECTROMETRY | 6 | 80% | 1% | 4 | Search CROSS+LINKING+MASS+SPECTROMETRY | Search CROSS+LINKING+MASS+SPECTROMETRY |
| 2 | CHEMICAL CROSS LINKING | 4 | 14% | 8% | 27 | Search CHEMICAL+CROSS+LINKING | Search CHEMICAL+CROSS+LINKING |
| 3 | CROSS LINKED PEPTIDES | 4 | 75% | 1% | 3 | Search CROSS+LINKED+PEPTIDES | Search CROSS+LINKED+PEPTIDES |
| 4 | IN VIVO PROTEIN INTERACTIONS | 2 | 67% | 1% | 2 | Search IN+VIVO+PROTEIN+INTERACTIONS | Search IN+VIVO+PROTEIN+INTERACTIONS |
| 5 | PROTEIN 3D STRUCTURE | 2 | 25% | 2% | 6 | Search PROTEIN+3D+STRUCTURE | Search PROTEIN+3D+STRUCTURE |
| 6 | PROTEIN INTERACTION REPORTER | 1 | 100% | 1% | 2 | Search PROTEIN+INTERACTION+REPORTER | Search PROTEIN+INTERACTION+REPORTER |
| 7 | XL MS | 1 | 100% | 1% | 2 | Search XL+MS | Search XL+MS |
| 8 | GCAP 2 | 1 | 33% | 1% | 2 | Search GCAP+2 | Search GCAP+2 |
| 9 | BIFUNCTIONAL CROSSLINKING | 1 | 50% | 0% | 1 | Search BIFUNCTIONAL+CROSSLINKING | Search BIFUNCTIONAL+CROSSLINKING |
| 10 | EDMAN CHEMISTRY | 1 | 50% | 0% | 1 | Search EDMAN+CHEMISTRY | Search EDMAN+CHEMISTRY |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | LINKED PEPTIDES | 51 | 56% | 19% | 63 |
| 2 | LINKING REAGENTS | 51 | 91% | 6% | 21 |
| 3 | UNIVERSAL ISOLATION | 6 | 100% | 1% | 4 |
| 4 | ALLOW IDENTIFICATION | 4 | 75% | 1% | 3 |
| 5 | PROTEIN INTERACTION ANALYSIS | 4 | 75% | 1% | 3 |
| 6 | TOP DOWN APPROACH | 4 | 17% | 6% | 19 |
| 7 | INTERACTION REPORTER TECHNOLOGY | 3 | 100% | 1% | 3 |
| 8 | LINKING ANALYSIS | 2 | 44% | 1% | 4 |
| 9 | ASP PRO BOND | 2 | 67% | 1% | 2 |
| 10 | MAPPING PROTEIN INTERFACES | 2 | 67% | 1% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Probing Native Protein Structures by Chemical Cross-linking, Mass Spectrometry, and Bioinformatics | 2010 | 144 | 79 | 46% |
| CROSSLINKING COMBINED WITH MASS SPECTROMETRY FOR STRUCTURAL PROTEOMICS | 2010 | 64 | 46 | 87% |
| Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions | 2006 | 252 | 89 | 43% |
| Chemical cross-linkers for protein structure studies by mass spectrometry | 2013 | 20 | 96 | 52% |
| Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry | 2010 | 31 | 40 | 58% |
| The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks | 2014 | 2 | 60 | 73% |
| Chemical Cross-linking and Mass Spectrometry for the Structural Analysis of Protein Assemblies | 2013 | 5 | 84 | 73% |
| In vivo protein complex topologies: Sights through a cross-linking lens | 2012 | 20 | 66 | 48% |
| Mass spectrometric analysis of cross-linking sites for the structure of proteins and protein complexes | 2008 | 41 | 50 | 56% |
| Modern Mass Spectrometry-Based Structural Proteomics | 2014 | 1 | 30 | 73% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PHARMACEUT CHEM BIOANALYT | 14 | 48% | 6.4% | 21 |
| 2 | GENOME BRITISH COLUMBIA PROTE | 1 | 16% | 2.4% | 8 |
| 3 | ADV SOFTWARE RD | 1 | 50% | 0.3% | 1 |
| 4 | BIODEF PROTEOM | 1 | 50% | 0.3% | 1 |
| 5 | GRP ARTIFICIAL BINDING PROT | 1 | 50% | 0.3% | 1 |
| 6 | MOL RUMENTAT | 1 | 50% | 0.3% | 1 |
| 7 | PHARMACEUT ANAL 1 | 1 | 50% | 0.3% | 1 |
| 8 | GENOME BRITISH COLUMBIA PROT | 1 | 22% | 0.6% | 2 |
| 9 | BIOMOL SYNTH GRP | 0 | 33% | 0.3% | 1 |
| 10 | CLEVELAND MASS SPE OMETRY IL | 0 | 33% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000183528 | TANDEM AFFINITY PURIFICATION//TAP TANDEM AFFINITY PURIFICATION//TANDEM AFFINITY PURIFICATION TAP |
| 2 | 0.0000145266 | HYDROGEN EXCHANGE//HYDROGEN DEUTERIUM EXCHANGE//AMIDE HYDROGEN EXCHANGE |
| 3 | 0.0000143272 | DIAZIRINE//PHOTOAFFINITY LABELING//BIORECOGNIT CHEM |
| 4 | 0.0000142823 | ION ION REACTIONS//ELECTRON CAPTURE DISSOCIATION//ELECTRON TRANSFER DISSOCIATION |
| 5 | 0.0000106638 | NON COVALENT COMPLEXES//NONCOVALENT COMPLEX//NONCOVALENT COMPLEXES |
| 6 | 0.0000081411 | PEPTIDE IDENTIFICATION//JOURNAL OF PROTEOME RESEARCH//QUANTITATIVE PROTEOMICS |
| 7 | 0.0000067171 | PEPTIDE FRAGMENTATION//COMPUTAT PROTE GRP//PROLINE EFFECT |
| 8 | 0.0000059705 | AFFINITY MASS SPECTROMETRY//PEPTIDE CHIP ANALYSIS//PROTEOLYTIC EPITOPE EXCISION |
| 9 | 0.0000038685 | SAMPLE PREFRACTIONATION//PREFRACTIONATION//ALKALINE PROTEINS |
| 10 | 0.0000037783 | BIMOLECULAR FLUORESCENCE COMPLEMENTATION//SPLIT UBIQUITIN//BIMOLECULAR FLUORESCENCE COMPLEMENTATION BIFC |