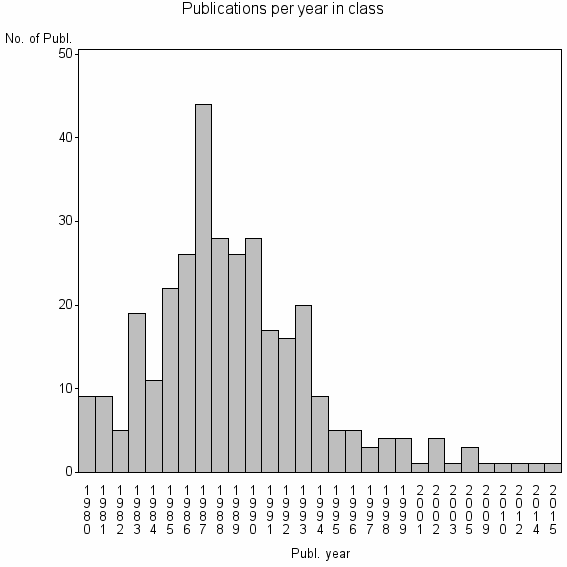
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22452 | 324 | 18.7 | 57% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1339 | 7890 | BIOTECHNIQUES//PCR-METHODS AND APPLICATIONS//DIFFERENTIAL DISPLAY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SPI SELECTION | Author keyword | 1 | 50% | 1% | 2 |
| 2 | CO PRECIPITANT | Author keyword | 1 | 50% | 0% | 1 |
| 3 | INSERT SIZE | Author keyword | 1 | 50% | 0% | 1 |
| 4 | PHAGE LAMBDA VECTORS | Author keyword | 1 | 50% | 0% | 1 |
| 5 | WISTAR RAT BRAIN | Author keyword | 1 | 50% | 0% | 1 |
| 6 | GENOMIC LIBRARIES | Author keyword | 1 | 18% | 1% | 3 |
| 7 | ARTHROBACTER AUREUS | Author keyword | 0 | 33% | 0% | 1 |
| 8 | CELL ASSOCIATED PROTEIN | Author keyword | 0 | 33% | 0% | 1 |
| 9 | CRE LOX SITE SPECIFIC RECOMBINATION | Author keyword | 0 | 33% | 0% | 1 |
| 10 | DROP DIALYSIS | Author keyword | 0 | 33% | 0% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SPI SELECTION | 1 | 50% | 1% | 2 | Search SPI+SELECTION | Search SPI+SELECTION |
| 2 | CO PRECIPITANT | 1 | 50% | 0% | 1 | Search CO+PRECIPITANT | Search CO+PRECIPITANT |
| 3 | INSERT SIZE | 1 | 50% | 0% | 1 | Search INSERT+SIZE | Search INSERT+SIZE |
| 4 | PHAGE LAMBDA VECTORS | 1 | 50% | 0% | 1 | Search PHAGE+LAMBDA+VECTORS | Search PHAGE+LAMBDA+VECTORS |
| 5 | WISTAR RAT BRAIN | 1 | 50% | 0% | 1 | Search WISTAR+RAT+BRAIN | Search WISTAR+RAT+BRAIN |
| 6 | GENOMIC LIBRARIES | 1 | 18% | 1% | 3 | Search GENOMIC+LIBRARIES | Search GENOMIC+LIBRARIES |
| 7 | ARTHROBACTER AUREUS | 0 | 33% | 0% | 1 | Search ARTHROBACTER+AUREUS | Search ARTHROBACTER+AUREUS |
| 8 | CELL ASSOCIATED PROTEIN | 0 | 33% | 0% | 1 | Search CELL+ASSOCIATED+PROTEIN | Search CELL+ASSOCIATED+PROTEIN |
| 9 | CRE LOX SITE SPECIFIC RECOMBINATION | 0 | 33% | 0% | 1 | Search CRE+LOX+SITE+SPECIFIC+RECOMBINATION | Search CRE+LOX+SITE+SPECIFIC+RECOMBINATION |
| 10 | DROP DIALYSIS | 0 | 33% | 0% | 1 | Search DROP+DIALYSIS | Search DROP+DIALYSIS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DIRECTIONAL CLONING | 1 | 50% | 1% | 2 |
| 2 | PHAGE PLASMID RECOMBINATION | 1 | 100% | 1% | 2 |
| 3 | DROSOPHILA CHROMATIN | 1 | 50% | 0% | 1 |
| 4 | LAMBDA VECTOR | 1 | 50% | 0% | 1 |
| 5 | PHOSPHATE PRECIPITATE FORMATION | 1 | 50% | 0% | 1 |
| 6 | BACTERIOPHAGE LIBRARIES | 1 | 29% | 1% | 2 |
| 7 | RECOMBINATION INVIVO | 0 | 33% | 0% | 1 |
| 8 | CDNA EXPRESSION VECTOR | 0 | 17% | 0% | 1 |
| 9 | EXPRESSION LIBRARIES | 0 | 13% | 0% | 1 |
| 10 | RARE TRANSCRIPTS | 0 | 11% | 0% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| BACTERIOPHAGE-LAMBDA AS A CLONING VECTOR | 1992 | 16 | 63 | 35% |
| Bacteriophage lambda-based expression vectors | 2001 | 7 | 11 | 45% |
| GENE ISOLATION BY SCREENING LAMBDA-GT11 LIBRARIES WITH ANTIBODIES | 1987 | 186 | 5 | 40% |
| 1ST-STRAND CDNA SYNTHESIS PRIMED WITH OLIGO(DT) | 1987 | 225 | 3 | 33% |
| CONSTRUCTION OF COMPLEX DIRECTIONAL COMPLEMENTARY-DNA LIBRARIES IN SFII | 1992 | 3 | 10 | 60% |
| DIRECT SEQUENCING OF LAMBDA-GT11 CLONES | 1993 | 2 | 17 | 59% |
| 2ND-STRAND CDNA SYNTHESIS - MESSENGER-RNA FRAGMENTS AS PRIMERS | 1987 | 58 | 3 | 67% |
| AMPLIFICATION, STORAGE, AND REPLICATION OF LIBRARIES | 1987 | 38 | 4 | 75% |
| 2ND-STRAND CDNA SYNTHESIS - CLASSICAL METHOD | 1987 | 15 | 2 | 100% |
| GENE ISOLATION WITH GAMMA-GT11 SYSTEM | 1991 | 9 | 16 | 31% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT MOL ECOPHYSIOL GRP | 0 | 33% | 0.3% | 1 |
| 2 | CIENCIAS BIOMED MICROBIOL | 0 | 25% | 0.3% | 1 |
| 3 | SERV GEN OYO INVEST | 0 | 14% | 0.3% | 1 |
| 4 | ENERGY CONVERS MAT TEAM | 0 | 100% | 0.3% | 1 |
| 5 | FSBISP BIOTECHNOL | 0 | 100% | 0.3% | 1 |
| 6 | IMMUNOL IMM18 10666 N TORREY PINES RD | 0 | 100% | 0.3% | 1 |
| 7 | MED COMMITTEE CELL DEV BIOL | 0 | 100% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000124104 | HN1//CANC 6401 E THOMAS RD//HORMONAL PROFILING |
| 2 | 0.0000109505 | POLYLINKER//STREPTOMYCIN SENSITIVITY//INSERTIONAL INACTIVATION |
| 3 | 0.0000091034 | EXPT BIOTECHNOL PLANT//HUMAN CONSENSUS INTERFERON ALPHA//A21 DESAMIDOINSULIN |
| 4 | 0.0000090351 | PLASMID PURIFICATION//SUPERCOILED PLASMID DNA//PLASMID DNA |
| 5 | 0.0000089379 | RBM5//HUMAN CHROMOSOME 3//LUCA 15 |
| 6 | 0.0000083372 | DYE TERMINATOR//ENERGY TRANSFER DYES//GENOMAT |
| 7 | 0.0000082745 | PIP92//LYMPHOCYTE PROTEINS//PROTEINPAEDIA |
| 8 | 0.0000078472 | EMANUEL SYNDROME//PARTIAL TRISOMY 11Q//DUPLICATION 11Q |
| 9 | 0.0000070793 | COMMON HAMSTER//GREATER LONG TAILED HAMSTER//CRICETUS CRICETUS |
| 10 | 0.0000066498 | PRIMULA NUTANS//INFECT DIS CLUSTER ADV MED DENT AMDI//SEASHORE MEADOW |