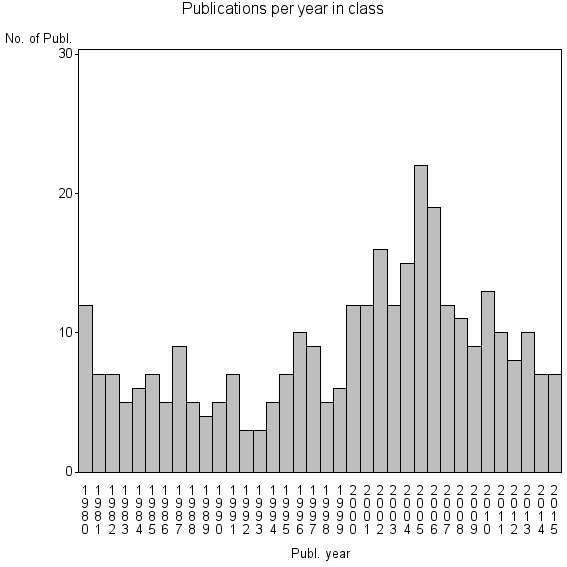
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22506 | 322 | 26.7 | 66% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1198 | 8823 | ASYMMETRIC REDUCTION//BAEYER VILLIGER OXIDATION//OMEGA TRANSAMINASE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | FRUCTOSYL AMINO ACID OXIDASE | Author keyword | 38 | 89% | 5% | 17 |
| 2 | FRUCTOSYL AMINE OXIDASE | Author keyword | 33 | 100% | 4% | 13 |
| 3 | FRUCTOSYL PEPTIDE OXIDASE | Author keyword | 23 | 100% | 3% | 10 |
| 4 | FRUCTOSAMINE 3 KINASE | Author keyword | 15 | 77% | 3% | 10 |
| 5 | DEGLYCATION | Author keyword | 12 | 75% | 3% | 9 |
| 6 | SARCOSINE OXIDASE | Author keyword | 8 | 42% | 4% | 14 |
| 7 | AMADORIASE | Author keyword | 5 | 63% | 2% | 5 |
| 8 | DIMETHYLGLYCINE DEHYDROGENASE | Author keyword | 5 | 63% | 2% | 5 |
| 9 | FN3K | Author keyword | 4 | 75% | 1% | 3 |
| 10 | FRUCTOSYL VALINE | Author keyword | 4 | 56% | 2% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | FRUCTOSYL AMINO ACID OXIDASE | 38 | 89% | 5% | 17 | Search FRUCTOSYL+AMINO+ACID+OXIDASE | Search FRUCTOSYL+AMINO+ACID+OXIDASE |
| 2 | FRUCTOSYL AMINE OXIDASE | 33 | 100% | 4% | 13 | Search FRUCTOSYL+AMINE+OXIDASE | Search FRUCTOSYL+AMINE+OXIDASE |
| 3 | FRUCTOSYL PEPTIDE OXIDASE | 23 | 100% | 3% | 10 | Search FRUCTOSYL+PEPTIDE+OXIDASE | Search FRUCTOSYL+PEPTIDE+OXIDASE |
| 4 | FRUCTOSAMINE 3 KINASE | 15 | 77% | 3% | 10 | Search FRUCTOSAMINE+3+KINASE | Search FRUCTOSAMINE+3+KINASE |
| 5 | DEGLYCATION | 12 | 75% | 3% | 9 | Search DEGLYCATION | Search DEGLYCATION |
| 6 | SARCOSINE OXIDASE | 8 | 42% | 4% | 14 | Search SARCOSINE+OXIDASE | Search SARCOSINE+OXIDASE |
| 7 | AMADORIASE | 5 | 63% | 2% | 5 | Search AMADORIASE | Search AMADORIASE |
| 8 | DIMETHYLGLYCINE DEHYDROGENASE | 5 | 63% | 2% | 5 | Search DIMETHYLGLYCINE+DEHYDROGENASE | Search DIMETHYLGLYCINE+DEHYDROGENASE |
| 9 | FN3K | 4 | 75% | 1% | 3 | Search FN3K | Search FN3K |
| 10 | FRUCTOSYL VALINE | 4 | 56% | 2% | 5 | Search FRUCTOSYL+VALINE | Search FRUCTOSYL+VALINE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EC 153 | 50 | 88% | 7% | 23 |
| 2 | N METHYLTRYPTOPHAN OXIDASE | 25 | 70% | 7% | 21 |
| 3 | SP SOIL STRAIN | 23 | 86% | 4% | 12 |
| 4 | CORYNEBACTERIUM SP 2 4 1 | 21 | 90% | 3% | 9 |
| 5 | AMADORIASE | 12 | 86% | 2% | 6 |
| 6 | FRUCTOSAMINE 3 KINASE RELATED PROTEIN | 12 | 86% | 2% | 6 |
| 7 | MONOMERIC SARCOSINE OXIDASE | 9 | 33% | 7% | 22 |
| 8 | DEGLYCATING ENZYMES | 8 | 100% | 2% | 5 |
| 9 | ASPERGILLUS SP | 7 | 26% | 7% | 24 |
| 10 | SARCOSINE DEHYDROGENASE | 6 | 45% | 3% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Enzymatic repair of Amadori products | 2012 | 10 | 39 | 62% |
| Occurrence, characteristics, and applications of fructosyl amine oxidases (amadoriases) | 2010 | 4 | 47 | 77% |
| Enzymatic deglycation of Amadori products in bacteria: mechanisms, occurrence and physiological functions | 2011 | 8 | 34 | 50% |
| Enzymatic deglycation of proteins | 2003 | 36 | 56 | 32% |
| The human flavoproteome | 2013 | 9 | 75 | 8% |
| Development of creatinine-degrading enzymes for application to clinical assays | 2001 | 0 | 16 | 81% |
| SARCOSINE OXIDASE - STRUCTURE, FUNCTION, AND THE APPLICATION TO CREATININE DETERMINATION | 1994 | 11 | 43 | 58% |
| NON-ENZYMATIC GLYCOSYLATION AND DEGLYCATING ENZYMES | 2010 | 6 | 69 | 19% |
| Novel diflavin-containing dehydrogenase family: FAD, FMN and ATP-containing L-proline dehydrogenase | 2006 | 1 | 15 | 33% |
| DEVELOPMENT OF BACTERIOPHAGE VECTORS FOR THE INDUSTRIAL USE AND THEIR APPLICATION TO THE PRODUCTION OF DIAGNOSTIC ENZYMES | 1991 | 1 | 5 | 80% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TECHNOL RISK MANAGEMENT | 1 | 38% | 0.9% | 3 |
| 2 | HORMONES METAB UNIT | 1 | 40% | 0.6% | 2 |
| 3 | AFDELING RONTGENDIAGNOSE | 1 | 50% | 0.3% | 1 |
| 4 | PHARMACEUT RD PROGRAM | 0 | 33% | 0.3% | 1 |
| 5 | FUNDAMENTAL LIFE SCI | 0 | 12% | 0.9% | 3 |
| 6 | CENT YODOGAWA KU | 0 | 11% | 0.9% | 3 |
| 7 | BIOMED NMR EENHEID | 0 | 25% | 0.3% | 1 |
| 8 | DETO SURG | 0 | 25% | 0.3% | 1 |
| 9 | HIGH THROUGHPUT TORY | 0 | 14% | 0.3% | 1 |
| 10 | ROUMEN TSANEV MOL BIOL | 0 | 14% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000198191 | 1 5 ANHYDRO D FRUCTOSE//1 5 ANHYDRO D GLUCITOL//PYRANOSE DEHYDROGENASE |
| 2 | 0.0000140270 | JAFFE REACTION//CREATININE ANALYSIS//CREATININE DEIMINASE |
| 3 | 0.0000095101 | FUMARATE REDUCTASE//COMPLEX II//WOLINELLA SUCCINOGENES |
| 4 | 0.0000086948 | NICOTINE DEGRADATION//NICOTINE CATABOLISM//PSEUDOMONAS SP HF 1 |
| 5 | 0.0000085643 | ADVANCED GLYCATION END PRODUCTS//GLYCATION//PENTOSIDINE |
| 6 | 0.0000085134 | P HYDROXYBENZOATE HYDROXYLASE//3 HYDROXYBENZOATE 6 HYDROXYLASE//4 AMINOBENZOATE HYDROXYLASE |
| 7 | 0.0000082409 | HBA1C//GLYCATED ALBUMIN//GLYCATED HEMOGLOBIN |
| 8 | 0.0000072885 | SERINE HYDROXYMETHYLTRANSFERASE//THREONINE ALDOLASE//BIOCATALYT CHEM |
| 9 | 0.0000067589 | HYDANTOINASE//D HYDANTOINASE//L N CARBAMOYLASE |
| 10 | 0.0000061635 | STREPTOMYCES ANSOCHROMOGENES//POLYOXAMIC ACID//NIKKOMYCIN BIOSYNTHESIS |