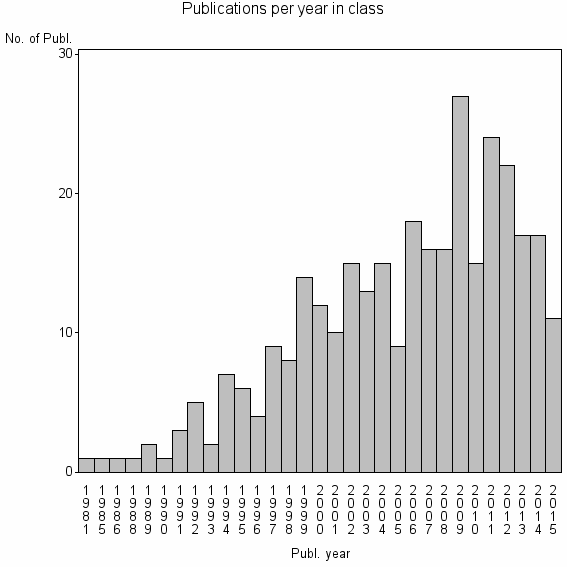
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22524 | 322 | 40.8 | 89% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | OBG | Author keyword | 23 | 100% | 3% | 10 |
| 2 | DRG2 | Author keyword | 8 | 100% | 2% | 5 |
| 3 | BACTERIAL GTPASE | Author keyword | 6 | 100% | 1% | 4 |
| 4 | HFLX | Author keyword | 6 | 100% | 1% | 4 |
| 5 | YJEQ | Author keyword | 6 | 100% | 1% | 4 |
| 6 | YSXC | Author keyword | 6 | 100% | 1% | 4 |
| 7 | CGTA | Author keyword | 4 | 75% | 1% | 3 |
| 8 | ENGA | Author keyword | 4 | 50% | 2% | 6 |
| 9 | CGTA PROTEIN | Author keyword | 3 | 100% | 1% | 3 |
| 10 | YGGG GENE | Author keyword | 3 | 100% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | OBG | 23 | 100% | 3% | 10 | Search OBG | Search OBG |
| 2 | DRG2 | 8 | 100% | 2% | 5 | Search DRG2 | Search DRG2 |
| 3 | BACTERIAL GTPASE | 6 | 100% | 1% | 4 | Search BACTERIAL+GTPASE | Search BACTERIAL+GTPASE |
| 4 | HFLX | 6 | 100% | 1% | 4 | Search HFLX | Search HFLX |
| 5 | YJEQ | 6 | 100% | 1% | 4 | Search YJEQ | Search YJEQ |
| 6 | YSXC | 6 | 100% | 1% | 4 | Search YSXC | Search YSXC |
| 7 | CGTA | 4 | 75% | 1% | 3 | Search CGTA | Search CGTA |
| 8 | ENGA | 4 | 50% | 2% | 6 | Search ENGA | Search ENGA |
| 9 | CGTA PROTEIN | 3 | 100% | 1% | 3 | Search CGTA+PROTEIN | Search CGTA+PROTEIN |
| 10 | YGGG GENE | 3 | 100% | 1% | 3 | Search YGGG+GENE | Search YGGG+GENE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | OBG | 80 | 100% | 8% | 25 |
| 2 | P LOOP GTPASES | 23 | 86% | 4% | 12 |
| 3 | BACTERIAL GTPASES | 23 | 76% | 5% | 16 |
| 4 | MEMBRANE PROTEIN FEOB | 21 | 90% | 3% | 9 |
| 5 | RNC OPERON | 21 | 85% | 3% | 11 |
| 6 | BINDING PROTEIN OBG | 19 | 71% | 5% | 15 |
| 7 | INSERTIONAL MUTANT | 15 | 88% | 2% | 7 |
| 8 | FERROUS IRON TRANSPORTER | 13 | 80% | 2% | 8 |
| 9 | YJEQ | 12 | 86% | 2% | 6 |
| 10 | YLQF | 12 | 86% | 2% | 6 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis | 2014 | 5 | 83 | 65% |
| Role of GTPases in Bacterial Ribosome Assembly | 2009 | 50 | 95 | 56% |
| The Universally Conserved Prokaryotic GTPases | 2011 | 34 | 284 | 48% |
| Function of the universally conserved bacterial GTPases | 2003 | 78 | 23 | 74% |
| GTPases involved in bacterial ribosome maturation | 2013 | 2 | 110 | 85% |
| Structure-Function Relationships of the G Domain, a Canonical Switch Motif | 2011 | 88 | 165 | 16% |
| The weird and wonderful world of bacterial ribosome regulation | 2007 | 91 | 208 | 23% |
| The Obg subfamily of bacterial GTP-binding proteins: essential proteins of largely unknown functions that are evolutionarily conserved from bacteria to humans | 2005 | 17 | 42 | 81% |
| Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function | 2001 | 109 | 70 | 33% |
| Comparative genomics of prokaryotic GTP-binding proteins (the Era, Obg, EngA, ThdF (TrmE), YchF and YihA families) and their relationship to eukaryotic GTP-binding proteins (the DRG, ARF, RAB, RAN, RAS and RHO families) | 2001 | 38 | 74 | 42% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CENT GENOME | 1 | 38% | 0.9% | 3 |
| 2 | BK21 EB NCRC | 1 | 50% | 0.3% | 1 |
| 3 | CNRS BIOPHYS MOL CELLULAIRE | 1 | 50% | 0.3% | 1 |
| 4 | HUAIHE RIVER HOSP | 1 | 50% | 0.3% | 1 |
| 5 | IGBMCEQUIPE ELLISEE LA LIGUEUMR 7104U964 | 1 | 50% | 0.3% | 1 |
| 6 | LIFE SCI STRUCT BIOL | 1 | 50% | 0.3% | 1 |
| 7 | MICROBIAL CELL BIOL | 1 | 50% | 0.3% | 1 |
| 8 | ALBERTA RNA TRAINING | 1 | 29% | 0.6% | 2 |
| 9 | CELLULAR MOL BIOLMINATO KU | 0 | 12% | 0.9% | 3 |
| 10 | HEMATOL ONCOL INFECT DIS | 0 | 17% | 0.6% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000192894 | NUCLEOSTEMIN//GNL3L//ALKEK BIOSCI TECHNOL |
| 2 | 0.0000113693 | ESSENTIAL GENES//SUBTRACTIVE GENOMICS//ESSENTIAL GENE |
| 3 | 0.0000107001 | DAP3//ICT1//MITOCHONDRIAL RIBOSOMAL PROTEINS |
| 4 | 0.0000105838 | FARNESYLTHIOSALICYLIC ACID//SALIRASIB//ABT STRUKTURELLE BIOL |
| 5 | 0.0000093088 | PPGPP//STRINGENT RESPONSE//RPOS |
| 6 | 0.0000090700 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 7 | 0.0000087571 | MRNA ANALOG//80S RIBOSOME//HUMAN RIBOSOME |
| 8 | 0.0000076305 | QUEUINE//TRNA MODIFICATION//RNA MODIFICATION |
| 9 | 0.0000065984 | RNASE E//RNASE G//POLYNUCLEOTIDE PHOSPHORYLASE |
| 10 | 0.0000063451 | TID1//HRFI//DNAJB1 |