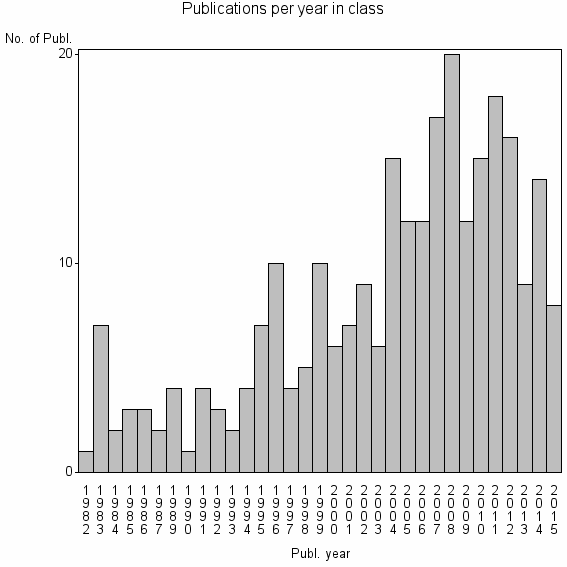
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 24177 | 268 | 41.6 | 82% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 201 | 20439 | MELAS//MITOCHONDRIAL DNA//LEBERS HEREDITARY OPTIC NEUROPATHY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | NUMT | Author keyword | 40 | 62% | 15% | 41 |
| 2 | NUMTS | Author keyword | 22 | 56% | 10% | 27 |
| 3 | NUMTDNA | Author keyword | 6 | 100% | 1% | 4 |
| 4 | NUCLEAR INSERTION | Author keyword | 4 | 75% | 1% | 3 |
| 5 | NUCLEAR MITOCHONDRIAL PSEUDOGENES | Author keyword | 4 | 75% | 1% | 3 |
| 6 | NUPT | Author keyword | 3 | 57% | 1% | 4 |
| 7 | NUPTS | Author keyword | 3 | 57% | 1% | 4 |
| 8 | MITOCHONDRIAL PSEUDOGENES | Author keyword | 3 | 60% | 1% | 3 |
| 9 | NUCLEAR INTEGRATION | Author keyword | 2 | 67% | 1% | 2 |
| 10 | NUCLEAR MITOCHONDRIAL PSEUDOGENE | Author keyword | 2 | 67% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NUMT | 40 | 62% | 15% | 41 | Search NUMT | Search NUMT |
| 2 | NUMTS | 22 | 56% | 10% | 27 | Search NUMTS | Search NUMTS |
| 3 | NUMTDNA | 6 | 100% | 1% | 4 | Search NUMTDNA | Search NUMTDNA |
| 4 | NUCLEAR INSERTION | 4 | 75% | 1% | 3 | Search NUCLEAR+INSERTION | Search NUCLEAR+INSERTION |
| 5 | NUCLEAR MITOCHONDRIAL PSEUDOGENES | 4 | 75% | 1% | 3 | Search NUCLEAR+MITOCHONDRIAL+PSEUDOGENES | Search NUCLEAR+MITOCHONDRIAL+PSEUDOGENES |
| 6 | NUPT | 3 | 57% | 1% | 4 | Search NUPT | Search NUPT |
| 7 | NUPTS | 3 | 57% | 1% | 4 | Search NUPTS | Search NUPTS |
| 8 | MITOCHONDRIAL PSEUDOGENES | 3 | 60% | 1% | 3 | Search MITOCHONDRIAL+PSEUDOGENES | Search MITOCHONDRIAL+PSEUDOGENES |
| 9 | NUCLEAR INTEGRATION | 2 | 67% | 1% | 2 | Search NUCLEAR+INTEGRATION | Search NUCLEAR+INTEGRATION |
| 10 | NUCLEAR MITOCHONDRIAL PSEUDOGENE | 2 | 67% | 1% | 2 | Search NUCLEAR+MITOCHONDRIAL+PSEUDOGENE | Search NUCLEAR+MITOCHONDRIAL+PSEUDOGENE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NUMTS | 25 | 70% | 8% | 21 |
| 2 | HUMAN NUCLEAR GENOME | 15 | 82% | 3% | 9 |
| 3 | HUMAN MITOCHONDRIAL PSEUDOGENES | 14 | 64% | 5% | 14 |
| 4 | DNA LIKE SEQUENCES | 12 | 75% | 3% | 9 |
| 5 | CORRESPONDING NUCLEAR PSEUDOGENE | 6 | 100% | 1% | 4 |
| 6 | NUCLEAR GENOME | 5 | 17% | 11% | 29 |
| 7 | CYTOCHROME B PSEUDOGENE | 4 | 75% | 1% | 3 |
| 8 | NUMT | 3 | 50% | 1% | 4 |
| 9 | NUCLEAR PSEUDOGENES | 2 | 67% | 1% | 2 |
| 10 | ORGANELLE DNA | 2 | 21% | 3% | 9 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Molecular Poltergeists: Mitochondrial DNA Copies (numts) in Sequenced Nuclear Genomes | 2010 | 116 | 83 | 59% |
| Mitochondrial pseudogenes: evolution's misplaced witnesses | 2001 | 545 | 54 | 65% |
| Nuclear integrations: Challenges for mitochondrial DNA markers | 1996 | 557 | 28 | 50% |
| DNA Transfer from Organelles to the Nucleus: The Idiosyncratic Genetics of Endosymbiosis | 2009 | 102 | 135 | 27% |
| Reconstructing evolution: gene transfer from plastids to the nucleus | 2008 | 57 | 53 | 30% |
| Origin, evolution and genetic effects of nuclear insertions of organelle DNA | 2005 | 78 | 68 | 50% |
| Pseudomitochondrial genome haunts disease studies | 2008 | 52 | 34 | 26% |
| ROLE OF INTERCOMPARTMENTAL DNA TRANSFER IN PRODUCING GENETIC DIVERSITY | 2011 | 8 | 198 | 26% |
| Nuclear mitochondrial pseudogenes | 2010 | 6 | 109 | 34% |
| Organellar genes - why do they end up in the nucleus? | 2000 | 88 | 50 | 16% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CANC INCA SYNERGIE | 1 | 50% | 0.4% | 1 |
| 2 | CIRM LEVU MICROBIOL GENET MOL | 1 | 50% | 0.4% | 1 |
| 3 | DEV METABOL NEUROL BRANCH | 1 | 50% | 0.4% | 1 |
| 4 | FREDERICK CANC DEV GENOM ERS | 1 | 50% | 0.4% | 1 |
| 5 | INRA CNRS AGROPARISTECH | 1 | 50% | 0.4% | 1 |
| 6 | MATH UNDER PROGRAM | 1 | 50% | 0.4% | 1 |
| 7 | NUCLEO GENET LICADA CONSERVACAO BIO ERSIDADE | 1 | 50% | 0.4% | 1 |
| 8 | PL MARINE MAMMAL ECOL GRP | 1 | 50% | 0.4% | 1 |
| 9 | TROP FISHERIES AQUACULTURE | 1 | 50% | 0.4% | 1 |
| 10 | ABT PFLANZENZUCHTUNG GENET | 1 | 25% | 0.7% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000158490 | PROCESSED PSEUDOGENES//PHYSICAL AND PSYCHOLOGICAL STRESSES//DUPLICATED PSEUDOGENES |
| 2 | 0.0000122270 | CERCOPITHECINI//PAPIONINI//GENET INFORMAT PROGRAM |
| 3 | 0.0000119939 | DOUBLY UNIPARENTAL INHERITANCE//PATERNAL LEAKAGE//MITOCHONDRIAL POLARITY |
| 4 | 0.0000107923 | MITOCHONDRIAL DNA//MITOCHONDRIAL GENOME//MITOGENOME |
| 5 | 0.0000103766 | DNA BARCODING//BIO ERS ONTARIO//DNA BARCODE |
| 6 | 0.0000099540 | CYTOPLASMIC MALE STERILITY//RNA EDITING//CYTOPLASMIC MALE STERILITY CMS |
| 7 | 0.0000090361 | D310//MITOCHONDRIAL MICROSATELLITE INSTABILITY//MITOCHONDRIAL HAPLOGROUPS |
| 8 | 0.0000077073 | ENGLISH MEDICAL LAW//PROFESS ETH PEAK//WUNSCH UND WAHLRECHT |
| 9 | 0.0000075992 | NUCLEOMORPH//CHLORARACHNIOPHYTES//SECONDARY ENDOSYMBIOSIS |
| 10 | 0.0000073160 | ANCIENT DNA//GEOGENET//SKELETAL PREPARATION |