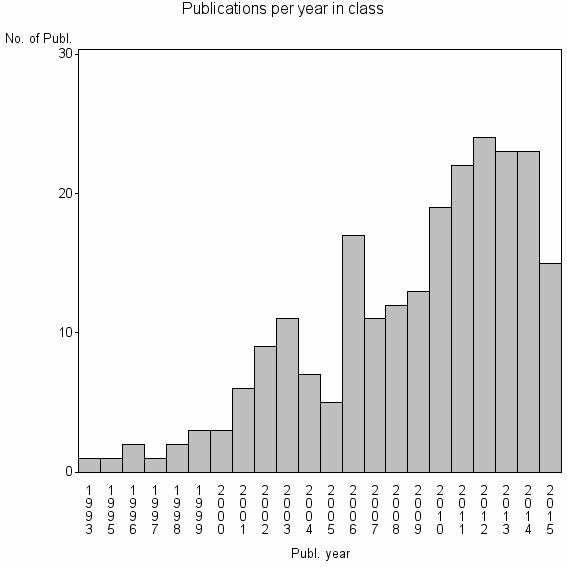
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 25566 | 230 | 51.9 | 93% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | U BOX | Author keyword | 7 | 53% | 4% | 10 |
| 2 | CELLULAR BIOL MOL GENET | Address | 4 | 75% | 1% | 3 |
| 3 | PLANT BIOL GRP PROGRAM | Address | 4 | 75% | 1% | 3 |
| 4 | RING E3 LIGASE | Author keyword | 3 | 57% | 2% | 4 |
| 5 | RING H2 | Author keyword | 3 | 43% | 3% | 6 |
| 6 | RING ZINC FINGER PROTEIN | Author keyword | 3 | 50% | 2% | 4 |
| 7 | ARM REPEAT | Author keyword | 3 | 60% | 1% | 3 |
| 8 | MAKORIN | Author keyword | 3 | 60% | 1% | 3 |
| 9 | E2 E3 INTERACTION | Author keyword | 2 | 67% | 1% | 2 |
| 10 | SAUL1 | Author keyword | 2 | 67% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | U BOX | 7 | 53% | 4% | 10 | Search U+BOX | Search U+BOX |
| 2 | RING E3 LIGASE | 3 | 57% | 2% | 4 | Search RING+E3+LIGASE | Search RING+E3+LIGASE |
| 3 | RING H2 | 3 | 43% | 3% | 6 | Search RING+H2 | Search RING+H2 |
| 4 | RING ZINC FINGER PROTEIN | 3 | 50% | 2% | 4 | Search RING+ZINC+FINGER+PROTEIN | Search RING+ZINC+FINGER+PROTEIN |
| 5 | ARM REPEAT | 3 | 60% | 1% | 3 | Search ARM+REPEAT | Search ARM+REPEAT |
| 6 | MAKORIN | 3 | 60% | 1% | 3 | Search MAKORIN | Search MAKORIN |
| 7 | E2 E3 INTERACTION | 2 | 67% | 1% | 2 | Search E2+E3+INTERACTION | Search E2+E3+INTERACTION |
| 8 | SAUL1 | 2 | 67% | 1% | 2 | Search SAUL1 | Search SAUL1 |
| 9 | SPL11 | 2 | 67% | 1% | 2 | Search SPL11 | Search SPL11 |
| 10 | ARTIFICIAL E3 | 1 | 100% | 1% | 2 | Search ARTIFICIAL+E3 | Search ARTIFICIAL+E3 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMBINATORY ROLES | 8 | 100% | 2% | 5 |
| 2 | ATAIRP1 | 3 | 100% | 1% | 3 |
| 3 | MEDIATED DROUGHT STRESS | 3 | 100% | 1% | 3 |
| 4 | FINGER E3 LIGASE | 3 | 42% | 2% | 5 |
| 5 | MKRN1 | 2 | 67% | 1% | 2 |
| 6 | RING H2 FINGER PROTEIN | 2 | 67% | 1% | 2 |
| 7 | L CV PUKANG | 2 | 50% | 1% | 3 |
| 8 | PROTEASOME PROTEOLYTIC PATHWAY | 2 | 11% | 6% | 13 |
| 9 | N ACETYLCHITOOLIGOSACCHARIDE ELICITOR | 1 | 12% | 5% | 12 |
| 10 | EL5 | 1 | 100% | 1% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling | 2014 | 10 | 94 | 30% |
| Abiotic stress tolerance mediated by protein ubiquitination | 2012 | 61 | 155 | 19% |
| ATLs and BTLs, plant-specific and general eukaryotic structurally-related E3 ubiquitin ligases | 2014 | 2 | 31 | 45% |
| Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis | 2011 | 31 | 86 | 20% |
| The ubiquitin-proteasome system: central modifier of plant signalling | 2012 | 41 | 135 | 12% |
| The ubiquitin-26S proteasome system at the nexus of plant biology | 2009 | 278 | 136 | 7% |
| Plant E3 Ligases: Flexible Enzymes in a Sessile World | 2013 | 7 | 203 | 15% |
| The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates | 2009 | 57 | 83 | 19% |
| Ubiquitin ligases mediate growth and development by promoting protein death | 2007 | 65 | 69 | 16% |
| The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins | 2003 | 275 | 69 | 9% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CELLULAR BIOL MOL GENET | 4 | 75% | 1.3% | 3 |
| 2 | PLANT BIOL GRP PROGRAM | 4 | 75% | 1.3% | 3 |
| 3 | PL PLANT SCI TECHNOL | 1 | 40% | 0.9% | 2 |
| 4 | PHARMACEUT HLTH CARE | 1 | 12% | 2.2% | 5 |
| 5 | HAINAN BANANA GENET IMPROVEMENT | 1 | 25% | 0.9% | 2 |
| 6 | EAT PROGRAM | 0 | 11% | 1.7% | 4 |
| 7 | ADV INTEGRATED ANAL INFECT DIS | 0 | 33% | 0.4% | 1 |
| 8 | CROP GENE ENGN HUNAN PROVINCE | 0 | 33% | 0.4% | 1 |
| 9 | SCOTTISH CELL SIGNALLING SCILLS | 0 | 33% | 0.4% | 1 |
| 10 | POSTHARVEST PHYSIOL TECHNOL TROP HORT P | 0 | 25% | 0.4% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000258748 | SNRK2//ABI3//ABA RECEPTOR |
| 2 | 0.0000194078 | COP9 SIGNALOSOME//NEDD8//NEDDYLATION |
| 3 | 0.0000193743 | DREB//AP2 ERF//GCC BOX |
| 4 | 0.0000184491 | DSK2//UBIQUILIN 1//LUBAC |
| 5 | 0.0000113578 | WRKY TRANSCRIPTION FACTOR//WRKY//WRKY PROTEIN |
| 6 | 0.0000097717 | TYPE III EFFECTOR//MOLECULAR PLANT-MICROBE INTERACTIONS//SGT1 |
| 7 | 0.0000077558 | POLYUBIQUITIN GENE//UBIQUITYL CALMODULIN SYNTHETASE//FREE UBIQUITIN |
| 8 | 0.0000076732 | ARGINYLATION//JOHANSON BLIZZARD SYNDROME//N END RULE |
| 9 | 0.0000069228 | TRIM5 ALPHA//TRIMCYP//TRIM5 |
| 10 | 0.0000066048 | MCPIP1//ZC3H12A//MCPIP |