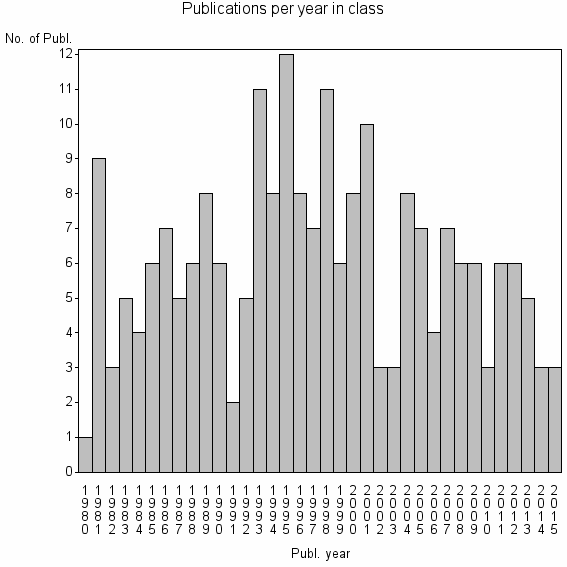
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 26064 | 218 | 35.6 | 60% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2915 | 2265 | GALACTOSEMIA//GALACTOSE 1 PHOSPHATE URIDYLTRANSFERASE//GALACTOSE 1 PHOSPHATE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | HISTIDINE BIOSYNTHESIS | Author keyword | 14 | 51% | 9% | 20 |
| 2 | IMIDAZOLEGLYCEROL PHOSPHATE DEHYDRATASE | Author keyword | 9 | 83% | 2% | 5 |
| 3 | HISTIDINOL DEHYDROGENASE | Author keyword | 7 | 53% | 4% | 9 |
| 4 | ATP PHOSPHORIBOSYLTRANSFERASE | Author keyword | 6 | 100% | 2% | 4 |
| 5 | OPERON EVOLUTION | Author keyword | 5 | 63% | 2% | 5 |
| 6 | IGP SYNTHASE | Author keyword | 3 | 60% | 1% | 3 |
| 7 | HIS3 | Author keyword | 2 | 38% | 2% | 5 |
| 8 | EVOLUTION OF METABOLIC PATHWAYS | Author keyword | 2 | 44% | 2% | 4 |
| 9 | IGPD | Author keyword | 2 | 67% | 1% | 2 |
| 10 | OPERON ORIGIN | Author keyword | 2 | 67% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HISTIDINE BIOSYNTHESIS | 14 | 51% | 9% | 20 | Search HISTIDINE+BIOSYNTHESIS | Search HISTIDINE+BIOSYNTHESIS |
| 2 | IMIDAZOLEGLYCEROL PHOSPHATE DEHYDRATASE | 9 | 83% | 2% | 5 | Search IMIDAZOLEGLYCEROL+PHOSPHATE+DEHYDRATASE | Search IMIDAZOLEGLYCEROL+PHOSPHATE+DEHYDRATASE |
| 3 | HISTIDINOL DEHYDROGENASE | 7 | 53% | 4% | 9 | Search HISTIDINOL+DEHYDROGENASE | Search HISTIDINOL+DEHYDROGENASE |
| 4 | ATP PHOSPHORIBOSYLTRANSFERASE | 6 | 100% | 2% | 4 | Search ATP+PHOSPHORIBOSYLTRANSFERASE | Search ATP+PHOSPHORIBOSYLTRANSFERASE |
| 5 | OPERON EVOLUTION | 5 | 63% | 2% | 5 | Search OPERON+EVOLUTION | Search OPERON+EVOLUTION |
| 6 | IGP SYNTHASE | 3 | 60% | 1% | 3 | Search IGP+SYNTHASE | Search IGP+SYNTHASE |
| 7 | HIS3 | 2 | 38% | 2% | 5 | Search HIS3 | Search HIS3 |
| 8 | EVOLUTION OF METABOLIC PATHWAYS | 2 | 44% | 2% | 4 | Search EVOLUTION+OF+METABOLIC+PATHWAYS | Search EVOLUTION+OF+METABOLIC+PATHWAYS |
| 9 | IGPD | 2 | 67% | 1% | 2 | Search IGPD | Search IGPD |
| 10 | OPERON ORIGIN | 2 | 67% | 1% | 2 | Search OPERON+ORIGIN | Search OPERON+ORIGIN |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | IMIDAZOLEGLYCEROLPHOSPHATE DEHYDRATASE | 11 | 100% | 3% | 6 |
| 2 | ADENOSINE TRIPHOSPHATE PHOSPHORIBOSYLTRANSFERASE | 8 | 100% | 2% | 5 |
| 3 | HISB GENE | 8 | 100% | 2% | 5 |
| 4 | DIRECTION SUBSTRATE KINETICS | 6 | 100% | 2% | 4 |
| 5 | HISTIDINE BIOSYNTHETIC GENES | 4 | 75% | 1% | 3 |
| 6 | IMIDAZOLEGLYCEROL PHOSPHATE DEHYDRATASE | 2 | 67% | 1% | 2 |
| 7 | HISTIDINE BIOSYNTHESIS | 2 | 24% | 4% | 8 |
| 8 | FIRST ENZYME | 2 | 50% | 1% | 3 |
| 9 | HISTIDINE BIOSYNTHETIC PATHWAY | 2 | 43% | 1% | 3 |
| 10 | GLUTAMINE AMIDOTRANSFERASE | 2 | 28% | 2% | 5 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Histidine biosynthesis in plants | 2006 | 30 | 49 | 43% |
| Targeting the Histidine Pathway in Mycobacterium tuberculosis | 2013 | 1 | 102 | 44% |
| Histidine biosynthetic pathway and genes: Structure, regulation, and evolution | 1996 | 122 | 160 | 31% |
| Histidine biosynthesis, its regulation and biotechnological application in Corynebacterium glutamicum | 2014 | 2 | 82 | 29% |
| Origin and evolution of metabolic pathways | 2009 | 24 | 79 | 18% |
| Zinc metalloenzymes as new targets against the bacterial pathogen Brucella | 2012 | 3 | 45 | 31% |
| REGULATION OF THE 1ST STEP OF THE HISTIDINE BIOSYNTHESIS IN ESCHERICHIA-COLI | 1988 | 5 | 1 | 100% |
| Contribution of structural genomics to understanding the biology of Escherichia coli | 2003 | 6 | 72 | 17% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DIPARTIMENTO BIOL CELLULARE SVILUPPO VIALE SCI | 1 | 100% | 0.9% | 2 |
| 2 | ECOLE SUPER CHIM MONTPELLIERIBMMUMR 5247 | 1 | 100% | 0.9% | 2 |
| 3 | MED 2 IST PATOL GEN CATTEDRA MICROBIOL | 1 | 100% | 0.9% | 2 |
| 4 | INT S | 1 | 20% | 2.3% | 5 |
| 5 | CNRSUMR 5236 | 1 | 40% | 0.9% | 2 |
| 6 | BIOCIENCIAS RIO CLARA | 1 | 50% | 0.5% | 1 |
| 7 | COMP CHEM UNIT | 1 | 50% | 0.5% | 1 |
| 8 | ECOLE SUPER CHIM MONTPELLIERIBMM | 1 | 50% | 0.5% | 1 |
| 9 | ENVIRONM FO TRY SCI | 1 | 50% | 0.5% | 1 |
| 10 | ETUD AGENTS PATHOGENES BIOTECHNOL SANTE CP | 1 | 50% | 0.5% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000170039 | UDP GLUCOSE DEHYDROGENASE//UDP GLCA//UDP GLUCURONATE |
| 2 | 0.0000125724 | TRYPTOPHAN SYNTHASE//INDOLE 3 GLYCEROL PHOSPHATE SYNTHASE//SECT ENZYME STRUCT FUNCT |
| 3 | 0.0000118788 | TERMINATION OF REPLICATION//REPLICATION TERMINATOR PROTEIN//FORK ARREST |
| 4 | 0.0000105083 | UDP SUGAR HYDROLASE//PHOSPHOHEXOMUTASE//HAD SUPERFAMILY |
| 5 | 0.0000085958 | PHOSPHORIBOSYLTRANSFERASE//MOL PARASITOL DRUG DESIGN//HGXPRT |
| 6 | 0.0000076341 | HOMOCITRATE SYNTHASE//ALPHA AMINOADIPATE REDUCTASE//ALPHA AMINOADIPATE PATHWAY |
| 7 | 0.0000068470 | GAL4P//GAL80//GAL GENES |
| 8 | 0.0000067195 | DIHYDROOROTASE//CARBAMOYL PHOSPHATE SYNTHETASE//ASPARTATE CARBAMOYLTRANSFERASE |
| 9 | 0.0000065155 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 10 | 0.0000061926 | NUSG//TRANSCRIPTION TERMINATION//RHO FACTOR |