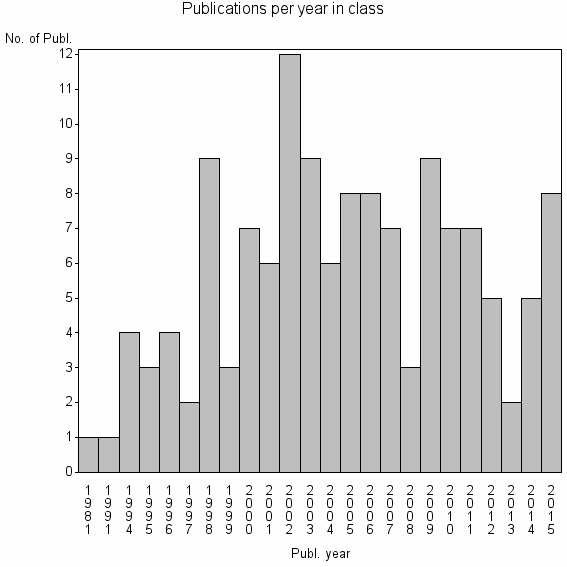
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 30576 | 136 | 24.6 | 63% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 524 | 14185 | ADAPTIVE DYNAMICS//EXPERIMENTAL EVOLUTION//PENNA MODEL |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | ERATO COMPLEX SYST BIOL | Address | 3 | 100% | 2% | 3 |
| 2 | ISOLOGOUS DIVERSIFICATION | Author keyword | 3 | 100% | 2% | 3 |
| 3 | COHERENT SPIKING | Author keyword | 2 | 67% | 1% | 2 |
| 4 | MINORITY CONTROL | Author keyword | 2 | 67% | 1% | 2 |
| 5 | REACTING KINETICS | Author keyword | 2 | 67% | 1% | 2 |
| 6 | COMPLEX SYST BIOL PROJECT | Address | 1 | 17% | 6% | 8 |
| 7 | BACTERIAL MUTANTS | Author keyword | 1 | 50% | 1% | 2 |
| 8 | KANEKO YOMO MODEL | Author keyword | 1 | 100% | 1% | 2 |
| 9 | MICROVOLUMES | Author keyword | 1 | 40% | 1% | 2 |
| 10 | MOLECULAR CHAOS | Author keyword | 1 | 33% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ISOLOGOUS DIVERSIFICATION | 3 | 100% | 2% | 3 | Search ISOLOGOUS+DIVERSIFICATION | Search ISOLOGOUS+DIVERSIFICATION |
| 2 | COHERENT SPIKING | 2 | 67% | 1% | 2 | Search COHERENT+SPIKING | Search COHERENT+SPIKING |
| 3 | MINORITY CONTROL | 2 | 67% | 1% | 2 | Search MINORITY+CONTROL | Search MINORITY+CONTROL |
| 4 | REACTING KINETICS | 2 | 67% | 1% | 2 | Search REACTING+KINETICS | Search REACTING+KINETICS |
| 5 | BACTERIAL MUTANTS | 1 | 50% | 1% | 2 | Search BACTERIAL+MUTANTS | Search BACTERIAL+MUTANTS |
| 6 | KANEKO YOMO MODEL | 1 | 100% | 1% | 2 | Search KANEKO+YOMO+MODEL | Search KANEKO+YOMO+MODEL |
| 7 | MICROVOLUMES | 1 | 40% | 1% | 2 | Search MICROVOLUMES | Search MICROVOLUMES |
| 8 | MOLECULAR CHAOS | 1 | 33% | 1% | 2 | Search MOLECULAR+CHAOS | Search MOLECULAR+CHAOS |
| 9 | BIOLOGICAL STATISTICAL MECHANICS | 1 | 50% | 1% | 1 | Search BIOLOGICAL+STATISTICAL+MECHANICS | Search BIOLOGICAL+STATISTICAL+MECHANICS |
| 10 | CELL TYPE DIVERSITY | 1 | 50% | 1% | 1 | Search CELL+TYPE+DIVERSITY | Search CELL+TYPE+DIVERSITY |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ISOLOGOUS DIVERSIFICATION | 16 | 58% | 13% | 18 |
| 2 | CELL SOCIETY | 15 | 68% | 10% | 13 |
| 3 | MOLECULAR TURNOVER CYCLES | 11 | 78% | 5% | 7 |
| 4 | MUTUAL SYNCHRONIZATION | 5 | 45% | 7% | 9 |
| 5 | MICROSCOPIC SELF ORGANIZATION | 3 | 40% | 4% | 6 |
| 6 | ALLOSTERIC ENZYMES | 3 | 30% | 5% | 7 |
| 7 | KINETIC ORIGIN | 2 | 67% | 1% | 2 |
| 8 | TURNOVER CYCLES | 2 | 67% | 1% | 2 |
| 9 | ROBUST DEVELOPMENT | 2 | 43% | 2% | 3 |
| 10 | MULTICELLULAR ORGANISMS | 1 | 19% | 4% | 6 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| ON RECURSIVE PRODUCTION AND EVOLVABILITY OF CELLS: CATALYTIC REACTION NETWORK APPROACH | 2005 | 7 | 26 | 42% |
| Microscopic self-organization in networks and regular lattice | 2002 | 0 | 12 | 58% |
| Identifying modifier genes of monogenic disease: strategies and difficulties | 2008 | 30 | 54 | 4% |
| The emergence of patterning in life's origin and evolution | 2009 | 5 | 51 | 4% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ERATO COMPLEX SYST BIOL | 3 | 100% | 2.2% | 3 |
| 2 | COMPLEX SYST BIOL PROJECT | 1 | 17% | 5.9% | 8 |
| 3 | IT CORE S | 1 | 50% | 0.7% | 1 |
| 4 | PROGRAMMING LANGUAGES | 1 | 50% | 0.7% | 1 |
| 5 | MICROB DIS | 0 | 33% | 0.7% | 1 |
| 6 | RECH EPIDEMIOL | 0 | 33% | 0.7% | 1 |
| 7 | UMR S794 | 0 | 33% | 0.7% | 1 |
| 8 | ERATO COMPLEX SYST BIOL PROJECT | 0 | 11% | 1.5% | 2 |
| 9 | FORM FUNCT PROJECT | 0 | 17% | 0.7% | 1 |
| 10 | INTELLIGENT COOPERAT CONTROL PROJECT | 0 | 17% | 0.7% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000189521 | COUPLED MAP LATTICES//INITIAL CONDITION ESTIMATION//GLOBALLY COUPLED MAPS |
| 2 | 0.0000148205 | LINEAR NOISE APPROXIMATION//STOCHASTIC GENE EXPRESSION//CHEMICAL MASTER EQUATION |
| 3 | 0.0000138154 | ROCK PAPER SCISSORS GAME//PAIR APPROXIMATION//ROCK SCISSORS PAPER GAME |
| 4 | 0.0000135186 | AVIDA//EVOLVABILITY//DIGITAL EVOLUTION |
| 5 | 0.0000095056 | RNA WORLD//ORIGIN OF LIFE//ORIGINS OF LIFE AND EVOLUTION OF BIOSPHERES |
| 6 | 0.0000085438 | ITO STRATONOVICH DILEMMA//INFORMAT ENGN COMP SCI MATH DISIM//AGO ANTAGONIST MODELS |
| 7 | 0.0000080237 | PROGRESSIVE STATE SELECTION//SPONTANEOUS TRANSFORMATION//GROWTH CONSTRAINT |
| 8 | 0.0000073378 | PROTOCELL//ELECTROFORMATION//MINIMAL CELL |
| 9 | 0.0000071008 | GLYCOLYTIC OSCILLATIONS//OSCILLATORY ENZYMES//NONLINEAR SYST BIOL |
| 10 | 0.0000069921 | COMPLEX QUANTITAT LINGUIST//MENZERATH ALTMANN LAW//FAMILY NAME DISTRIBUTION |