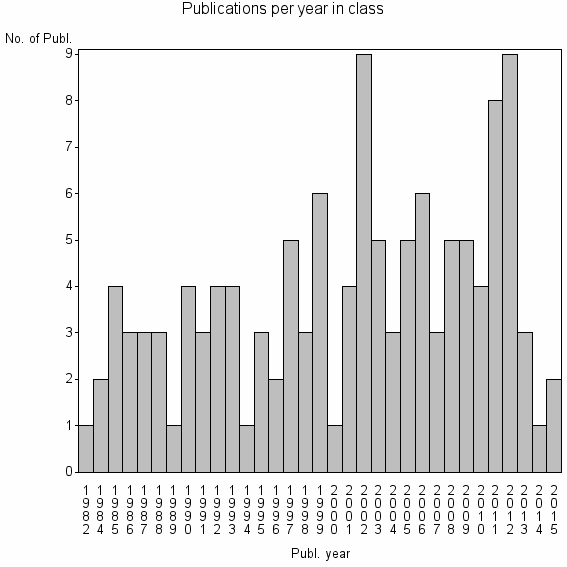
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 31254 | 125 | 36.3 | 81% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 501 | 14604 | RNA POLYMERASE//SITE SPECIFIC RECOMBINATION//H NS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EPIDEMIOLOGICAL SIGNIFICANCE | Author keyword | 2 | 67% | 2% | 2 |
| 2 | SIMILARITY IDENTIFICATION | Author keyword | 2 | 67% | 2% | 2 |
| 3 | AIRBORNE ESCHERICHIA COLI | Author keyword | 1 | 100% | 2% | 2 |
| 4 | MINIATURE INSERTION SEQUENCE | Author keyword | 1 | 100% | 2% | 2 |
| 5 | SINOGERMAN COOPERAT ZOONOSIS ANIM ORIGIN | Address | 1 | 100% | 2% | 2 |
| 6 | CHICKEN HOUSE | Author keyword | 1 | 40% | 2% | 2 |
| 7 | STEM LOOP STRUCTURES | Author keyword | 1 | 27% | 2% | 3 |
| 8 | ANIM DIS CONTROL ENGN | Address | 1 | 33% | 2% | 2 |
| 9 | BIME | Author keyword | 1 | 33% | 2% | 2 |
| 10 | MICROBIOLOGICAL DIAGNOSTIC | Author keyword | 1 | 33% | 2% | 2 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | REP SEQUENCES | 14 | 57% | 14% | 17 |
| 2 | INTERSPERSED MOSAIC ELEMENTS | 13 | 80% | 6% | 8 |
| 3 | EXTRAGENIC PALINDROMIC SEQUENCES | 6 | 27% | 15% | 19 |
| 4 | ERIC SEQUENCES | 4 | 67% | 3% | 4 |
| 5 | DNA INSERTION SEQUENCES | 3 | 60% | 2% | 3 |
| 6 | BIME | 2 | 67% | 2% | 2 |
| 7 | IS1397 | 2 | 67% | 2% | 2 |
| 8 | YERSINIAE | 2 | 50% | 2% | 3 |
| 9 | BIMES | 1 | 100% | 2% | 2 |
| 10 | NEISSERIAE | 1 | 50% | 2% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Miniature Repetitive Mobile Elements of Bacteria: Structural Organization and Properties | 2010 | 1 | 53 | 34% |
| SHORT, INTERSPERSED REPETITIVE DNA-SEQUENCES IN PROKARYOTIC GENOMES | 1992 | 208 | 69 | 26% |
| PALINDROMIC UNITS - A CASE OF HIGHLY REPETITIVE DNA-SEQUENCES IN BACTERIA | 1987 | 59 | 24 | 25% |
| BIME SEQUENCES - AN EXAMPLE OF ENTEROBACTERIAL REPETITIVE SEQUENCES | 1995 | 1 | 154 | 16% |
| The role of short sequence repeats in epidemiologic typing | 1999 | 47 | 59 | 3% |
| CONCEPT OF A BACTERIAL SPECIES | 1985 | 14 | 15 | 13% |
| Escherichia coli and Salmonella 2000: the view from here | 2001 | 16 | 55 | 2% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SINOGERMAN COOPERAT ZOONOSIS ANIM ORIGIN | 1 | 100% | 1.6% | 2 |
| 2 | ANIM DIS CONTROL ENGN | 1 | 33% | 1.6% | 2 |
| 3 | BIC GRANADA | 1 | 50% | 0.8% | 1 |
| 4 | URA 1444 | 1 | 17% | 2.4% | 3 |
| 5 | UNITE PROGRAMMAT MOL TOXICOL GENET | 0 | 10% | 3.2% | 4 |
| 6 | BIOMED ENGN PHYS SCI | 0 | 33% | 0.8% | 1 |
| 7 | OURCE SCI ENVIRONM ENGN | 0 | 33% | 0.8% | 1 |
| 8 | SEZIONE IGIENE | 0 | 33% | 0.8% | 1 |
| 9 | CNRS URA 1444 | 0 | 13% | 1.6% | 2 |
| 10 | MOL MICROBIAL ECOL ECOGENOM | 0 | 12% | 1.6% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000170668 | DNA TRANSPOSITION//PHAGE MU//TN10 |
| 2 | 0.0000144409 | RNASE E//RNASE G//POLYNUCLEOTIDE PHOSPHORYLASE |
| 3 | 0.0000119498 | EMANUEL SYNDROME//PARTIAL TRISOMY 11Q//DUPLICATION 11Q |
| 4 | 0.0000108409 | KAUFFMANN WHITE SCHEME//I CEUI//HMU UCFM INFECT GENOM |
| 5 | 0.0000101372 | OSCILLATORIALES//CHROOCOCCALES//PHYCOCYANIN OPERON |
| 6 | 0.0000084927 | TERMINATION OF REPLICATION//REPLICATION TERMINATOR PROTEIN//FORK ARREST |
| 7 | 0.0000081034 | H NS//HISTONE LIKE PROTEIN//BACTERIAL CHROMATIN |
| 8 | 0.0000074747 | NEISSERIA GONORRHOEAE//NEISSERIA MENINGITIDIS//LEHRSTUHL ZELLBIOL |
| 9 | 0.0000072153 | DNA GYRASE//GYRASE//REVERSE GYRASE |
| 10 | 0.0000069258 | SKIRBALL PROGRAM MOL PATHOGENESIS//BACTERIOPHAGE P2//RETRON |