Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
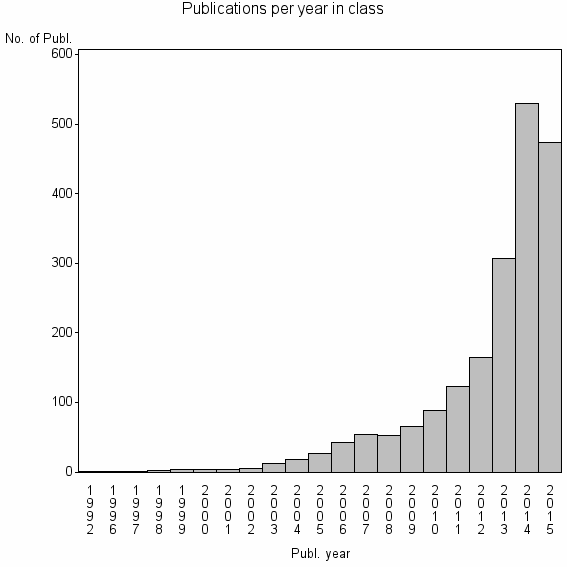
| 3138 | 1975 | 56.3 | 93% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | LONG NONCODING RNAS | Author keyword | 257 | 70% | 11% | 213 |
| 2 | LNCRNA | Author keyword | 180 | 68% | 8% | 156 |
| 3 | LONG NON CODING RNA | Author keyword | 157 | 71% | 6% | 126 |
| 4 | HOTAIR | Author keyword | 114 | 86% | 3% | 57 |
| 5 | LONG NON CODING RNAS | Author keyword | 77 | 76% | 3% | 54 |
| 6 | MALAT1 | Author keyword | 49 | 78% | 2% | 32 |
| 7 | LNCRNAS | Author keyword | 41 | 63% | 2% | 42 |
| 8 | LINCRNA | Author keyword | 38 | 70% | 2% | 31 |
| 9 | INCRNA | Author keyword | 34 | 68% | 2% | 30 |
| 10 | NON CODING RNA | Author keyword | 31 | 17% | 9% | 172 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LONG NONCODING RNAS | 257 | 70% | 11% | 213 | Search LONG+NONCODING+RNAS | Search LONG+NONCODING+RNAS |
| 2 | LNCRNA | 180 | 68% | 8% | 156 | Search LNCRNA | Search LNCRNA |
| 3 | LONG NON CODING RNA | 157 | 71% | 6% | 126 | Search LONG+NON+CODING+RNA | Search LONG+NON+CODING+RNA |
| 4 | HOTAIR | 114 | 86% | 3% | 57 | Search HOTAIR | Search HOTAIR |
| 5 | LONG NON CODING RNAS | 77 | 76% | 3% | 54 | Search LONG+NON+CODING+RNAS | Search LONG+NON+CODING+RNAS |
| 6 | MALAT1 | 49 | 78% | 2% | 32 | Search MALAT1 | Search MALAT1 |
| 7 | LNCRNAS | 41 | 63% | 2% | 42 | Search LNCRNAS | Search LNCRNAS |
| 8 | LINCRNA | 38 | 70% | 2% | 31 | Search LINCRNA | Search LINCRNA |
| 9 | INCRNA | 34 | 68% | 2% | 30 | Search INCRNA | Search INCRNA |
| 10 | NON CODING RNA | 31 | 17% | 9% | 172 | Search NON+CODING+RNA | Search NON+CODING+RNA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | HOTAIR | 178 | 94% | 3% | 63 |
| 2 | LONG NON CODING RNA | 118 | 48% | 9% | 181 |
| 3 | PROTEIN CODING RNA | 89 | 100% | 1% | 27 |
| 4 | PROSTATE SPECIFIC GENE | 79 | 87% | 2% | 39 |
| 5 | LONG NONCODING RNAS | 74 | 30% | 11% | 211 |
| 6 | INTEGRATIVE ANNOTATION | 72 | 91% | 2% | 30 |
| 7 | LINCRNAS | 38 | 76% | 1% | 26 |
| 8 | COMPETING ENDOGENOUS RNA | 31 | 92% | 1% | 12 |
| 9 | HULC | 27 | 92% | 1% | 11 |
| 10 | MALAT 1 | 27 | 83% | 1% | 15 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Long non-coding RNAs: new players in cell differentiation and development | 2014 | 123 | 123 | 61% |
| Long noncoding RNAs: Novel insights into gastric cancer | 2015 | 9 | 90 | 66% |
| Long Noncoding RNAs in Cardiovascular Diseases | 2015 | 9 | 110 | 42% |
| The Landscape of long noncoding RNA classification | 2015 | 6 | 153 | 50% |
| Long Noncoding RNAs: Cellular Address Codes in Development and Disease | 2013 | 190 | 99 | 58% |
| lincRNAs: Genomics, Evolution, and Mechanisms | 2013 | 200 | 174 | 53% |
| Long Noncoding RNAs and MicroRNAs in Cardiovascular Pathophysiology | 2015 | 7 | 130 | 37% |
| Long non-coding RNAs: insights into functions | 2009 | 865 | 47 | 57% |
| Genome Regulation by Long Noncoding RNAs | 2012 | 453 | 120 | 41% |
| Modular regulatory principles of large non-coding RNAs | 2012 | 299 | 95 | 52% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FUNCT RNOM TEAM | 10 | 73% | 0.4% | 8 |
| 2 | NINGBO YINZHOU PEOPLES HOSP | 8 | 75% | 0.3% | 6 |
| 3 | NEUROSCI NEUROL OPHTHALMOL | 6 | 80% | 0.2% | 4 |
| 4 | PROGRAM EPITHELIAL BIOL | 6 | 16% | 1.7% | 33 |
| 5 | AFFILIATED NANJING HOSP | 4 | 38% | 0.4% | 8 |
| 6 | HELMHOLTZ UNIV GRP MOL RNA BIOL CANC | 4 | 38% | 0.4% | 8 |
| 7 | TRANSLAT MED MATERNAL CHILDRENS HLTH | 3 | 100% | 0.2% | 3 |
| 8 | ARC SPECIAL FUNCT PL GENOM | 3 | 17% | 0.9% | 17 |
| 9 | NANJING MATERN CHILD HLTH CARE | 3 | 60% | 0.2% | 3 |
| 10 | PROGRAM INNATE IMMUN INFECT DIS IMMUNOL | 3 | 60% | 0.2% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000214612 | WRAP53//OVERLAPPING GENES//CUSTOM BIOTECHNOL SERV GRP |
| 2 | 0.0000208113 | SMALL ACTIVATING RNA//SARNA//RNA ACTIVATION |
| 3 | 0.0000142070 | P54NRB//POLYPYRIMIDINE TRACT BINDING PROTEIN ASSOCIATED SPLICING FACTOR//MATRIN 3 |
| 4 | 0.0000100073 | PROCESSED PSEUDOGENES//PHYSICAL AND PSYCHOLOGICAL STRESSES//DUPLICATED PSEUDOGENES |
| 5 | 0.0000090765 | CHIP SEQ//PEAK CALLING//DNASE SEQ |
| 6 | 0.0000084945 | RNA SEQ//DE NOVO ASSEMBLY//TRANSCRIPTOME ASSEMBLY |
| 7 | 0.0000080109 | XIST//X CHROMOSOME INACTIVATION//TSIX |
| 8 | 0.0000066554 | IROQUOIS//CONSERVED NONCODING ELEMENTS//ENERGY ENVIRONM BIOL COMP |
| 9 | 0.0000060486 | SPLICED LEADER//TRANS SPLICING//CIRCULAR RNA |
| 10 | 0.0000054261 | GENOMIC IMPRINTING//H19//BECKWITH WIEDEMANN SYNDROME |