Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
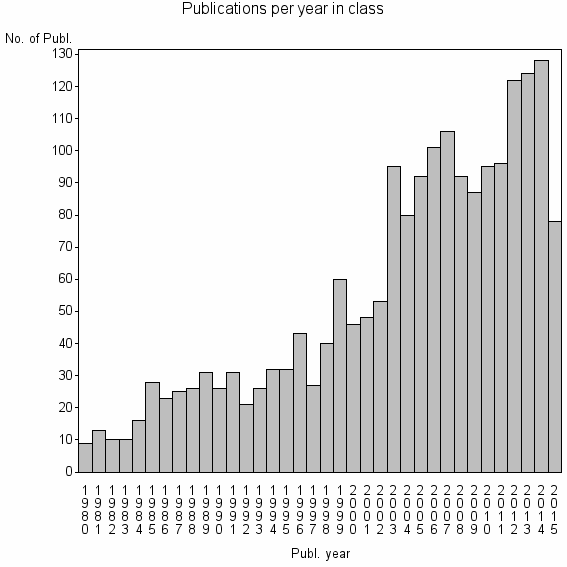
| 3151 | 1972 | 40.1 | 82% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TRANSLATIONAL SELECTION | Author keyword | 93 | 87% | 2% | 46 |
| 2 | CODON USAGE | Author keyword | 91 | 35% | 11% | 212 |
| 3 | SYNONYMOUS CODON USAGE | Author keyword | 88 | 80% | 3% | 55 |
| 4 | CODON USAGE BIAS | Author keyword | 73 | 54% | 5% | 94 |
| 5 | CODON BIAS | Author keyword | 34 | 36% | 4% | 77 |
| 6 | MUTATIONAL BIAS | Author keyword | 32 | 55% | 2% | 40 |
| 7 | MUTATIONAL PRESSURE | Author keyword | 27 | 78% | 1% | 18 |
| 8 | ORG EVOLUC GENOMA | Address | 26 | 61% | 1% | 28 |
| 9 | OPTIMAL CODONS | Author keyword | 26 | 87% | 1% | 13 |
| 10 | TRNA ABUNDANCE | Author keyword | 23 | 86% | 1% | 12 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRANSLATIONAL SELECTION | 93 | 87% | 2% | 46 | Search TRANSLATIONAL+SELECTION | Search TRANSLATIONAL+SELECTION |
| 2 | CODON USAGE | 91 | 35% | 11% | 212 | Search CODON+USAGE | Search CODON+USAGE |
| 3 | SYNONYMOUS CODON USAGE | 88 | 80% | 3% | 55 | Search SYNONYMOUS+CODON+USAGE | Search SYNONYMOUS+CODON+USAGE |
| 4 | CODON USAGE BIAS | 73 | 54% | 5% | 94 | Search CODON+USAGE+BIAS | Search CODON+USAGE+BIAS |
| 5 | CODON BIAS | 34 | 36% | 4% | 77 | Search CODON+BIAS | Search CODON+BIAS |
| 6 | MUTATIONAL BIAS | 32 | 55% | 2% | 40 | Search MUTATIONAL+BIAS | Search MUTATIONAL+BIAS |
| 7 | MUTATIONAL PRESSURE | 27 | 78% | 1% | 18 | Search MUTATIONAL+PRESSURE | Search MUTATIONAL+PRESSURE |
| 8 | OPTIMAL CODONS | 26 | 87% | 1% | 13 | Search OPTIMAL+CODONS | Search OPTIMAL+CODONS |
| 9 | TRNA ABUNDANCE | 23 | 86% | 1% | 12 | Search TRNA+ABUNDANCE | Search TRNA+ABUNDANCE |
| 10 | AMINO ACID USAGE | 21 | 64% | 1% | 21 | Search AMINO+ACID+USAGE | Search AMINO+ACID+USAGE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TRANSLATIONAL SELECTION | 304 | 84% | 9% | 168 |
| 2 | RESPECTIVE CODONS | 185 | 71% | 8% | 151 |
| 3 | MUTATIONAL BIASES | 147 | 92% | 3% | 58 |
| 4 | USAGE BIAS | 125 | 74% | 5% | 93 |
| 5 | UNICELLULAR ORGANISMS | 73 | 71% | 3% | 59 |
| 6 | ADAPTATION INDEX | 57 | 69% | 2% | 49 |
| 7 | SYNONYMOUS CODON USAGE | 50 | 35% | 6% | 117 |
| 8 | AMINO ACID USAGE | 48 | 65% | 2% | 46 |
| 9 | TRANSFER RNA ABUNDANCE | 46 | 71% | 2% | 37 |
| 10 | COLI TRANSFER RNAS | 39 | 61% | 2% | 41 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Synonymous but not the same: the causes and consequences of codon bias | 2011 | 253 | 131 | 66% |
| Selection on Codon Bias | 2008 | 216 | 45 | 64% |
| Determinants of translation efficiency and accuracy | 2011 | 97 | 123 | 59% |
| Speeding with control: codon usage, tRNAs, and ribosomes | 2012 | 49 | 77 | 58% |
| Hearing silence: non-neutral evolution at synonymous sites in mammals | 2006 | 370 | 112 | 32% |
| Understanding the contribution of synonymous mutations to human disease | 2011 | 165 | 86 | 29% |
| Codon bias and heterologous protein expression | 2004 | 399 | 69 | 30% |
| Codon usage: Nature's roadmap to expression and folding of proteins | 2011 | 41 | 97 | 62% |
| Engineering genes for predictable protein expression | 2012 | 24 | 71 | 52% |
| Genomic GC level, optimal growth temperature, and genome size in prokaryotes | 2006 | 47 | 7 | 86% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ORG EVOLUC GENOMA | 26 | 61% | 1.4% | 28 |
| 2 | DISTRIBUTED INFORMAT | 3 | 33% | 0.4% | 7 |
| 3 | EVOLUZ MOL | 3 | 22% | 0.6% | 11 |
| 4 | ESCUELA UNIV TECNOL MED | 3 | 60% | 0.2% | 3 |
| 5 | SUBTROP SERICULTURE MULBERRY OURCES PROTECT | 2 | 67% | 0.1% | 2 |
| 6 | CHEMINFORMAT BIOINFORMAT | 2 | 50% | 0.2% | 3 |
| 7 | O GENET MEJORA ANIM | 2 | 50% | 0.2% | 3 |
| 8 | ATELIER BIOINFORMAT | 2 | 20% | 0.5% | 9 |
| 9 | SECC BIOQUIM | 2 | 13% | 0.7% | 14 |
| 10 | FOOT MOUTH DIS REFERENCE | 2 | 15% | 0.6% | 11 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000164836 | ISOCHORES//EVOLUZ MOL//ISOCHORE |
| 2 | 0.0000106332 | TRIGGER FACTOR//NASCENT CHAIN//COTRANSLATIONAL FOLDING |
| 3 | 0.0000106156 | GENETIC CODE//EQUIPE BIOINFORMAT THEOR//CIRCULAR CODE |
| 4 | 0.0000102711 | FRAMESHIFTING//RIBOSOMAL FRAMESHIFTING//RNA PSEUDOKNOT |
| 5 | 0.0000100529 | SD SEQUENCE//RIBOSOMAL PROTEIN S1//SHINE DALGARNO SEQUENCE |
| 6 | 0.0000098597 | LATENT PERIODICITY//3 BASE PERIODICITY//DNA WALK |
| 7 | 0.0000064811 | SEX BIASED GENE EXPRESSION//SEXUAL ANTAGONISM//INTRALOCUS SEXUAL CONFLICT |
| 8 | 0.0000060293 | TREE OF LIFE//HORIZONTAL GENE TRANSFER//LATERAL GENE TRANSFER |
| 9 | 0.0000053770 | Q BIT//PAIR PREFERENCES//AMINO ACID DISTANCE MATRIX |
| 10 | 0.0000045901 | GRAPHICAL REPRESENTATION//ALIGNMENT FREE//SIMILARITIES DISSIMILARITIES |