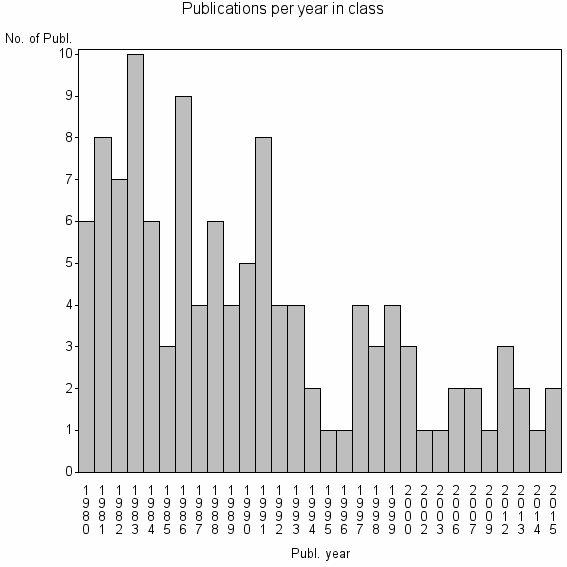
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 31849 | 117 | 32.0 | 44% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2150 | 4533 | 3 ISOPROPYLMALATE DEHYDROGENASE//GLUTAMATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SUCCINYL COA SYNTHETASE | Author keyword | 4 | 34% | 9% | 10 |
| 2 | OFF VP | Address | 2 | 36% | 4% | 5 |
| 3 | CANADA GRP PROT STRUCT FUNCT | Address | 1 | 38% | 3% | 3 |
| 4 | PHOSPHORYLATED HISTIDINE | Author keyword | 1 | 100% | 2% | 2 |
| 5 | NETWORK ADAPTABILITY | Author keyword | 1 | 50% | 1% | 1 |
| 6 | PHOSPHOENZYME FORMATION | Author keyword | 1 | 50% | 1% | 1 |
| 7 | SUCCINYL COA LIGASE | Author keyword | 1 | 29% | 2% | 2 |
| 8 | SUBSTRATE LEVEL PHOSPHORYLATION | Author keyword | 1 | 22% | 2% | 2 |
| 9 | MTA ELTE N MOL SYST NEUROBIOL | Address | 0 | 33% | 1% | 1 |
| 10 | SE NEUROBIOCHEM | Address | 0 | 33% | 1% | 1 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | THIOKINASE | 7 | 56% | 8% | 9 |
| 2 | SUBSTRATE SYNERGISM | 2 | 67% | 2% | 2 |
| 3 | THIOKINASES | 2 | 50% | 3% | 3 |
| 4 | 3 SULFINOPROPIONIC ACID | 1 | 50% | 1% | 1 |
| 5 | GTP SYNTHESIS | 0 | 33% | 1% | 1 |
| 6 | MIMIGARDEFORDENSIS STRAIN DPN7T | 0 | 25% | 1% | 1 |
| 7 | CATALYTIC COOPERATIVITY | 0 | 20% | 1% | 1 |
| 8 | ADVENELLA MIMIGARDEFORDENSIS | 0 | 14% | 1% | 1 |
| 9 | GLUTAMATE DEHYDROGENASES | 0 | 14% | 1% | 1 |
| 10 | 3 3 DITHIODIPROPIONIC ACID | 0 | 11% | 1% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| SUCCINYL-COA SYNTHETASE STRUCTURE-FUNCTION-RELATIONSHIPS AND OTHER CONSIDERATIONS | 1986 | 40 | 29 | 76% |
| CONTRIBUTION OF SUBUNIT INTERACTIONS TO THE EFFECTIVENESS OF CATALYSIS BY SUCCINYL COENZYME-A SYNTHETASE | 1984 | 4 | 10 | 70% |
| AN NMR PROBE OF SUCCINYL-COENZYME-A SYNTHETASE - SUBUNIT INTERACTIONS AND THE MECHANISM OF ACTION | 1983 | 10 | 22 | 41% |
| FLIP-FLOP MECHANISM IN THE ENZYMATIC CATALYSIS - MODEL FOR KINETIC DESCRIPTION OR PHYSICAL REALITY | 1990 | 0 | 37 | 8% |
| SUCCINIC THIOKINASE AND METABOLIC CONTROL | 1981 | 38 | 5 | 20% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | OFF VP | 2 | 36% | 4.3% | 5 |
| 2 | CANADA GRP PROT STRUCT FUNCT | 1 | 38% | 2.6% | 3 |
| 3 | MTA ELTE N MOL SYST NEUROBIOL | 0 | 33% | 0.9% | 1 |
| 4 | SE NEUROBIOCHEM | 0 | 33% | 0.9% | 1 |
| 5 | GENOM MED RARE DISORDERS | 0 | 11% | 0.9% | 1 |
| 6 | MTC CANADA | 0 | 100% | 0.9% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000285405 | 2 METHYLCITRIC ACID CYCLE//PROPIONATE CATABOLISM//PROPIONATE METABOLISM |
| 2 | 0.0000124314 | NM23//NUCLEOSIDE DIPHOSPHATE KINASE//NM23 H1 |
| 3 | 0.0000122334 | DISACCHARIDE NUCLEOSIDES//DISACCHARIDE NUCLEOSIDE//2 O MODIFIED NUCLEOSIDES |
| 4 | 0.0000102845 | POLG//MNGIE//DEOXYRIBONUCLEOSIDE KINASE |
| 5 | 0.0000101786 | ACETYL COA CARBOXYLASE//ACC ALPHA//4 PIPERIDINYL PIPERAZINE |
| 6 | 0.0000080771 | CYANELLES//CYANOPHORA PARADOXA//SILICONE OIL LAYER CENTRIFUGATION |
| 7 | 0.0000074244 | LIPID PHASE SEPARATIONS//SYNAPTOSOMAL MITOCHONDRIA//UNITE BIOMATH TOXICOL |
| 8 | 0.0000066914 | TRANSCRIPTION FACTOR TNRA//GLNR//BECKMAN COULTER |
| 9 | 0.0000061621 | GLYCERALDEHYDE 3 PHOSPHATE DEHYDROGENASE//D GLYCERALDEHYDE 3 PHOSPHATE DEHYDROGENASE//GRP CELLULAR NEUROBIOL |
| 10 | 0.0000054864 | 3 ISOPROPYLMALATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE//ISOPROPYLMALATE DEHYDROGENASE |