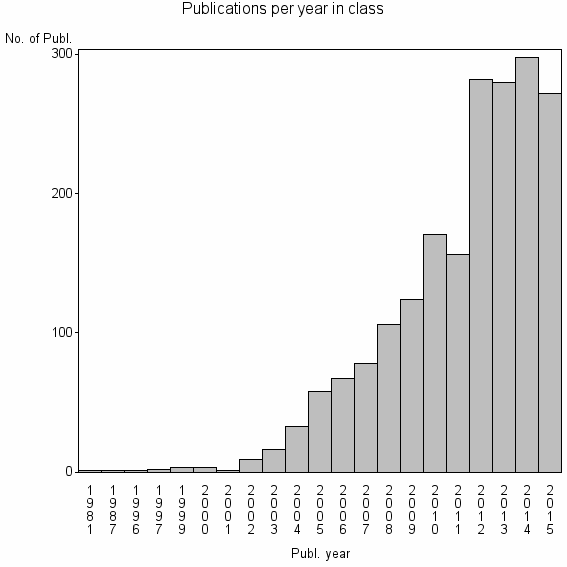
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 3202 | 1962 | 56.5 | 93% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 777 | 11593 | MADS BOX GENES//MADS BOX//FLOWER DEVELOPMENT |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MIR156 | Author keyword | 70 | 89% | 2% | 32 |
| 2 | DCL1 | Author keyword | 37 | 100% | 1% | 14 |
| 3 | MIR172 | Author keyword | 36 | 79% | 1% | 23 |
| 4 | TASIRNA | Author keyword | 29 | 88% | 1% | 14 |
| 5 | TRANS ACTING SIRNA | Author keyword | 24 | 91% | 1% | 10 |
| 6 | HYL1 | Author keyword | 21 | 90% | 0% | 9 |
| 7 | MIR164 | Author keyword | 21 | 85% | 1% | 11 |
| 8 | MIR159 | Author keyword | 20 | 100% | 0% | 9 |
| 9 | DEGRADOME | Author keyword | 17 | 41% | 2% | 31 |
| 10 | SBP BOX GENE | Author keyword | 14 | 100% | 0% | 7 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MIR156 | 70 | 89% | 2% | 32 | Search MIR156 | Search MIR156 |
| 2 | DCL1 | 37 | 100% | 1% | 14 | Search DCL1 | Search DCL1 |
| 3 | MIR172 | 36 | 79% | 1% | 23 | Search MIR172 | Search MIR172 |
| 4 | TASIRNA | 29 | 88% | 1% | 14 | Search TASIRNA | Search TASIRNA |
| 5 | TRANS ACTING SIRNA | 24 | 91% | 1% | 10 | Search TRANS+ACTING+SIRNA | Search TRANS+ACTING+SIRNA |
| 6 | HYL1 | 21 | 90% | 0% | 9 | Search HYL1 | Search HYL1 |
| 7 | MIR164 | 21 | 85% | 1% | 11 | Search MIR164 | Search MIR164 |
| 8 | MIR159 | 20 | 100% | 0% | 9 | Search MIR159 | Search MIR159 |
| 9 | DEGRADOME | 17 | 41% | 2% | 31 | Search DEGRADOME | Search DEGRADOME |
| 10 | SBP BOX GENE | 14 | 100% | 0% | 7 | Search SBP+BOX+GENE | Search SBP+BOX+GENE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT MICRORNAS | 798 | 84% | 22% | 429 |
| 2 | SMALL RNAS | 198 | 30% | 28% | 554 |
| 3 | CONSERVED MICRORNAS | 189 | 91% | 4% | 79 |
| 4 | TRANS ACTING SIRNAS | 180 | 58% | 11% | 207 |
| 5 | REGULATED MICRORNAS | 135 | 78% | 5% | 90 |
| 6 | BINDING PROTEIN HYL1 | 132 | 85% | 4% | 71 |
| 7 | PLANT MICRORNA | 128 | 94% | 2% | 46 |
| 8 | STRESS RESPONSIVE MICRORNAS | 117 | 89% | 3% | 54 |
| 9 | RESPONSIVE MICRORNAS | 114 | 88% | 3% | 53 |
| 10 | STRESS REGULATED MICRORNAS | 93 | 83% | 3% | 53 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| MicroRNA: a new target for improving plant tolerance to abiotic stress | 2015 | 6 | 102 | 71% |
| The Diversity, Biogenesis, and Activities of Endogenous Silencing Small RNAs in Arabidopsis | 2014 | 47 | 144 | 60% |
| MicroRNA-Based Biotechnology for Plant Improvement | 2015 | 5 | 233 | 70% |
| Classification and Comparison of Small RNAs from Plants | 2013 | 105 | 121 | 63% |
| Origin, Biogenesis, and Activity of Plant MicroRNAs | 2009 | 717 | 110 | 67% |
| Functions of microRNAs in plant stress responses | 2012 | 141 | 76 | 74% |
| Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs | 2013 | 80 | 211 | 55% |
| Evolution and Functional Diversification of MIRNA Genes | 2011 | 192 | 114 | 73% |
| The role of microRNAs in the control of flowering time | 2014 | 16 | 92 | 76% |
| Role of microRNAs in plant responses to nutrient stress | 2014 | 18 | 126 | 56% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | INTEGRAT GENOME BIOL | 9 | 15% | 2.8% | 55 |
| 2 | PLANT BIOL PHD PROGRAM | 9 | 83% | 0.3% | 5 |
| 3 | GENE SUPP S | 8 | 62% | 0.4% | 8 |
| 4 | CHEMGEN IGERT PROGRAM | 6 | 71% | 0.3% | 5 |
| 5 | MED PLANT CULTIVAT | 5 | 63% | 0.3% | 5 |
| 6 | FO T ECOL PROTECT | 4 | 47% | 0.4% | 7 |
| 7 | ALSON H SMITH AGR EXTENS | 4 | 75% | 0.2% | 3 |
| 8 | KEY IL CROP OURCES GENET IMPROVEMENT | 4 | 75% | 0.2% | 3 |
| 9 | OASIS ECOL AGR BINTUAN | 3 | 100% | 0.2% | 3 |
| 10 | CROP GENOM GENET IMPROVEMENT MOA | 3 | 33% | 0.4% | 8 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000159120 | RNA SILENCING//POST TRANSCRIPTIONAL GENE SILENCING//VIRUS INDUCED GENE SILENCING |
| 2 | 0.0000140841 | GW182//DROSHA//ARGONAUTE |
| 3 | 0.0000104197 | SHOOT APICAL MERISTEM//LEAF DEVELOPMENT//CLAVATA |
| 4 | 0.0000082933 | MSAP//RNA DIRECTED DNA METHYLATION//RDDM |
| 5 | 0.0000057309 | FLOWERING TIME//FLORIGEN//FLOWERING LOCUS T |
| 6 | 0.0000053536 | CIS MOTIF//ARABIDOPSIS T87 CELL LINE//GENE ATLAS |
| 7 | 0.0000050051 | PIRNA//PIWI//PIRNAS |
| 8 | 0.0000049240 | WRAP53//OVERLAPPING GENES//CUSTOM BIOTECHNOL SERV GRP |
| 9 | 0.0000047578 | AUXIN TRANSPORT//AUXIN//POLAR AUXIN TRANSPORT |
| 10 | 0.0000046956 | DREB//AP2 ERF//GCC BOX |