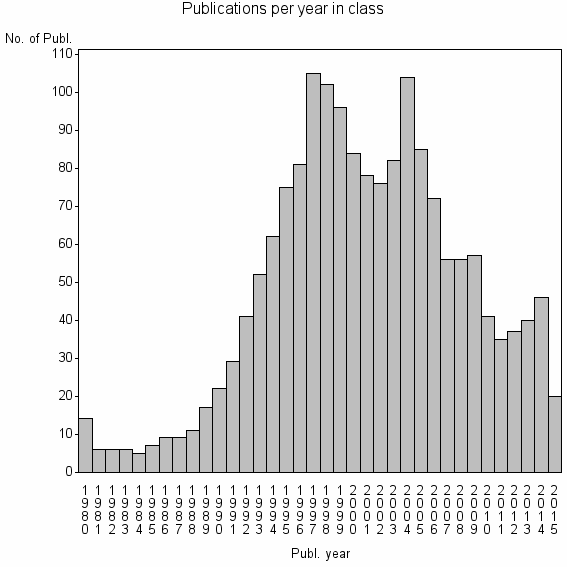
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 4359 | 1724 | 37.8 | 73% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | HP MODEL | Author keyword | 33 | 58% | 2% | 38 |
| 2 | STATISTICAL POTENTIALS | Author keyword | 18 | 52% | 1% | 25 |
| 3 | PROTEIN STRUCTURE PREDICTION | Author keyword | 17 | 14% | 7% | 120 |
| 4 | OFF LATTICE MODEL | Author keyword | 15 | 68% | 1% | 13 |
| 5 | KNOWLEDGE BASED POTENTIALS | Author keyword | 14 | 45% | 1% | 24 |
| 6 | LATTICE PROTEINS | Author keyword | 13 | 80% | 0% | 8 |
| 7 | KNOWLEDGE BASED POTENTIAL | Author keyword | 10 | 44% | 1% | 17 |
| 8 | THREADING | Author keyword | 9 | 19% | 3% | 44 |
| 9 | PROTEIN DESIGNABILITY | Author keyword | 9 | 83% | 0% | 5 |
| 10 | INVERSE FOLDING | Author keyword | 9 | 47% | 1% | 14 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HP MODEL | 33 | 58% | 2% | 38 | Search HP+MODEL | Search HP+MODEL |
| 2 | STATISTICAL POTENTIALS | 18 | 52% | 1% | 25 | Search STATISTICAL+POTENTIALS | Search STATISTICAL+POTENTIALS |
| 3 | PROTEIN STRUCTURE PREDICTION | 17 | 14% | 7% | 120 | Search PROTEIN+STRUCTURE+PREDICTION | Search PROTEIN+STRUCTURE+PREDICTION |
| 4 | OFF LATTICE MODEL | 15 | 68% | 1% | 13 | Search OFF+LATTICE+MODEL | Search OFF+LATTICE+MODEL |
| 5 | KNOWLEDGE BASED POTENTIALS | 14 | 45% | 1% | 24 | Search KNOWLEDGE+BASED+POTENTIALS | Search KNOWLEDGE+BASED+POTENTIALS |
| 6 | LATTICE PROTEINS | 13 | 80% | 0% | 8 | Search LATTICE+PROTEINS | Search LATTICE+PROTEINS |
| 7 | KNOWLEDGE BASED POTENTIAL | 10 | 44% | 1% | 17 | Search KNOWLEDGE+BASED+POTENTIAL | Search KNOWLEDGE+BASED+POTENTIAL |
| 8 | THREADING | 9 | 19% | 3% | 44 | Search THREADING | Search THREADING |
| 9 | PROTEIN DESIGNABILITY | 9 | 83% | 0% | 5 | Search PROTEIN+DESIGNABILITY | Search PROTEIN+DESIGNABILITY |
| 10 | INVERSE FOLDING | 9 | 47% | 1% | 14 | Search INVERSE+FOLDING | Search INVERSE+FOLDING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | QUASI CHEMICAL APPROXIMATION | 56 | 64% | 3% | 54 |
| 2 | KNOWLEDGE BASED POTENTIALS | 38 | 47% | 3% | 59 |
| 3 | ENERGY FUNCTIONS | 34 | 28% | 6% | 104 |
| 4 | CONTACT ENERGIES | 31 | 66% | 2% | 29 |
| 5 | HP MODEL | 27 | 63% | 2% | 27 |
| 6 | MEAN FORCE | 26 | 15% | 9% | 158 |
| 7 | DESIGNABILITY | 25 | 71% | 1% | 20 |
| 8 | STRUCTURE SELECTION | 23 | 38% | 3% | 48 |
| 9 | CONTACT POTENTIALS | 22 | 81% | 1% | 13 |
| 10 | GAS REFERENCE STATE | 20 | 52% | 2% | 28 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FOLDING & DESIGN | 3 | 12% | 1% | 22 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Protein Dynamism and Evolvability | 2009 | 293 | 44 | 14% |
| Effective energy functions for protein structure prediction | 2000 | 238 | 67 | 49% |
| In quest of an empirical potential for protein structure prediction | 2006 | 67 | 42 | 43% |
| Statistical potentials for fold assessment | 2002 | 142 | 99 | 61% |
| Inter-residue interactions in protein folding and stability | 2004 | 152 | 152 | 30% |
| Biophysics of protein evolution and evolutionary protein biophysics | 2014 | 10 | 416 | 17% |
| Ab initio protein structure prediction: Progress and prospects | 2001 | 132 | 72 | 42% |
| Reduced models of proteins and their applications | 2004 | 86 | 275 | 44% |
| Heteropolymer freezing and design: Towards physical models of protein folding | 2000 | 185 | 140 | 39% |
| The structure of protein evolution and the evolution of protein structure | 2008 | 37 | 76 | 41% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMPUTAT PROTEOM BIOPHYS | 8 | 60% | 0.5% | 9 |
| 2 | GSIT | 2 | 50% | 0.2% | 3 |
| 3 | LAURENCE H BAKER BIOINFORMAT BIOL STAT | 2 | 22% | 0.4% | 7 |
| 4 | FIS TEOR LICADA | 1 | 100% | 0.1% | 2 |
| 5 | OID SCI BIOMAT | 1 | 50% | 0.1% | 2 |
| 6 | PHYS ENGN TANDOGAN | 1 | 100% | 0.1% | 2 |
| 7 | THEORET BIOINFORMAT GRP | 1 | 100% | 0.1% | 2 |
| 8 | LH BAKER BIOINFORMAT BIOL STAT | 1 | 13% | 0.4% | 7 |
| 9 | MOL STRUCT SECT | 1 | 17% | 0.3% | 5 |
| 10 | VBI | 1 | 25% | 0.2% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000224346 | PROTEIN FOLDING//PHI VALUE//CONTACT ORDER |
| 2 | 0.0000186413 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//CASP |
| 3 | 0.0000178455 | SECONDARY STRUCTURE PREDICTION//PROTEIN SECONDARY STRUCTURE PREDICTION//COMPOUND PYRAMID MODEL |
| 4 | 0.0000171611 | COMPUTATIONAL PROTEIN DESIGN//DEAD END ELIMINATION//COMPUTATIONAL ENZYME DESIGN |
| 5 | 0.0000154812 | BAKER CHEM CHEM BIOL//REPLICA EXCHANGE METHOD//GENERALIZED ENSEMBLE ALGORITHM |
| 6 | 0.0000150818 | BIOINFORMAT TELEMED//HYDROPHOBICITY DEFICIENCY//HELIX INTERACTIONS |
| 7 | 0.0000134143 | LOOP PREDICTION//LOOP MODELING//STEPWISE FOLDING |
| 8 | 0.0000120209 | CORRELATED MUTATIONS//CONTACT PREDICTION//CONTACT MAP PREDICTION |
| 9 | 0.0000085544 | STAPHYLOCOCCAL NUCLEASE//THERMODYNAMIC MOLECULAR SWITCH//PROTEIN THERMOSTABILITY |
| 10 | 0.0000081699 | PINNING MODELS//HARRIS CRITERION//PINNING MODEL |