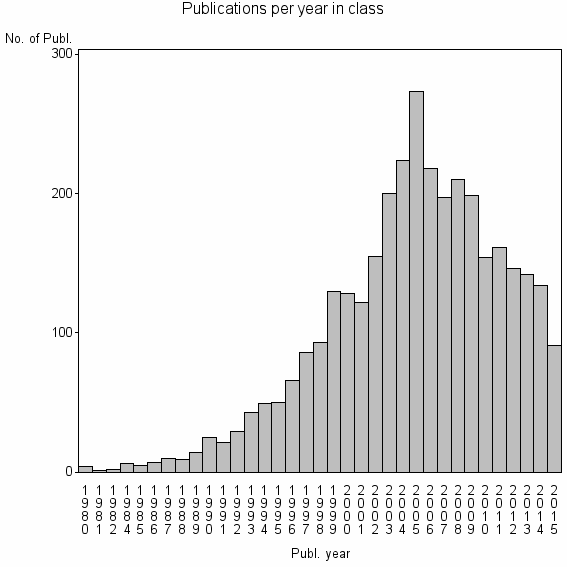
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 436 | 3404 | 37.0 | 78% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN STRUCTURE PREDICTION | Author keyword | 96 | 30% | 8% | 267 |
| 2 | FOLD RECOGNITION | Author keyword | 85 | 51% | 4% | 120 |
| 3 | CASP | Author keyword | 74 | 60% | 2% | 81 |
| 4 | PROTEIN STRUCTURE ALIGNMENT | Author keyword | 42 | 72% | 1% | 33 |
| 5 | CAFASP | Author keyword | 37 | 100% | 0% | 14 |
| 6 | PROTEIN STRUCTURE COMPARISON | Author keyword | 35 | 56% | 1% | 43 |
| 7 | THREADING | Author keyword | 33 | 34% | 2% | 80 |
| 8 | LIVEBENCH | Author keyword | 30 | 100% | 0% | 12 |
| 9 | STRUCTURE ALIGNMENT | Author keyword | 24 | 52% | 1% | 32 |
| 10 | PROTEIN THREADING | Author keyword | 24 | 56% | 1% | 29 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN STRUCTURE PREDICTION | 96 | 30% | 8% | 267 | Search PROTEIN+STRUCTURE+PREDICTION | Search PROTEIN+STRUCTURE+PREDICTION |
| 2 | FOLD RECOGNITION | 85 | 51% | 4% | 120 | Search FOLD+RECOGNITION | Search FOLD+RECOGNITION |
| 3 | CASP | 74 | 60% | 2% | 81 | Search CASP | Search CASP |
| 4 | PROTEIN STRUCTURE ALIGNMENT | 42 | 72% | 1% | 33 | Search PROTEIN+STRUCTURE+ALIGNMENT | Search PROTEIN+STRUCTURE+ALIGNMENT |
| 5 | CAFASP | 37 | 100% | 0% | 14 | Search CAFASP | Search CAFASP |
| 6 | PROTEIN STRUCTURE COMPARISON | 35 | 56% | 1% | 43 | Search PROTEIN+STRUCTURE+COMPARISON | Search PROTEIN+STRUCTURE+COMPARISON |
| 7 | THREADING | 33 | 34% | 2% | 80 | Search THREADING | Search THREADING |
| 8 | LIVEBENCH | 30 | 100% | 0% | 12 | Search LIVEBENCH | Search LIVEBENCH |
| 9 | STRUCTURE ALIGNMENT | 24 | 52% | 1% | 32 | Search STRUCTURE+ALIGNMENT | Search STRUCTURE+ALIGNMENT |
| 10 | PROTEIN THREADING | 24 | 56% | 1% | 29 | Search PROTEIN+THREADING | Search PROTEIN+THREADING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FOLD RECOGNITION | 189 | 44% | 10% | 324 |
| 2 | STRUCTURE ALIGNMENT | 173 | 54% | 6% | 220 |
| 3 | SCOP | 99 | 51% | 4% | 138 |
| 4 | PSI BLAST | 72 | 30% | 6% | 204 |
| 5 | MYCOPLASMA GENITALIUM PROTEINS | 69 | 83% | 1% | 39 |
| 6 | CATH | 65 | 62% | 2% | 68 |
| 7 | SEQUENCE PROFILES | 65 | 81% | 1% | 39 |
| 8 | FOLD SPACE | 47 | 64% | 1% | 46 |
| 9 | STRUCTURE PREDICTION | 44 | 11% | 11% | 368 |
| 10 | SUPERFAMILIES | 42 | 48% | 2% | 64 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre | 2008 | 276 | 40 | 73% |
| Progress and challenges in protein structure prediction | 2008 | 172 | 49 | 90% |
| Profile hidden Markov models | 1998 | 2356 | 60 | 48% |
| MODELLER: Generation and refinement of homology-based protein structure models | 2003 | 551 | 64 | 66% |
| Comparative modeling for protein structure prediction | 2006 | 142 | 41 | 98% |
| Advances and pitfalls of protein structural alignment | 2009 | 139 | 63 | 90% |
| A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction | 2005 | 204 | 28 | 86% |
| The impact of structural genomics: Expectations and outcomes | 2006 | 216 | 18 | 61% |
| Protein structure comparison: implications for the nature of 'fold space', and structure and function prediction | 2006 | 78 | 36 | 89% |
| Sequence comparison and protein structure prediction | 2006 | 91 | 103 | 78% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PL MOL ENGN | 17 | 55% | 0.6% | 21 |
| 2 | PRACT INFORMAT BIOINFORMAT GRP | 11 | 78% | 0.2% | 7 |
| 3 | PELS FAMILY BIOCHEM STRUCT BIOL | 9 | 50% | 0.4% | 13 |
| 4 | STUDY SYST BIOL | 8 | 30% | 0.7% | 23 |
| 5 | EBGM | 8 | 50% | 0.3% | 11 |
| 6 | BIOMOL STRUCT MODELLING UNIT | 7 | 32% | 0.6% | 19 |
| 7 | MED BIOL DIGITAL LIB | 6 | 80% | 0.1% | 4 |
| 8 | COMPUTAT BIOSCI SECT | 6 | 71% | 0.1% | 5 |
| 9 | EQUIPE BIOINFORMAT GENOM MOL | 6 | 31% | 0.4% | 15 |
| 10 | U726 | 5 | 41% | 0.3% | 9 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000256884 | SECONDARY STRUCTURE PREDICTION//PROTEIN SECONDARY STRUCTURE PREDICTION//COMPOUND PYRAMID MODEL |
| 2 | 0.0000219066 | COMPLEX SCI SOFTWARE//3D ZERNIKE DESCRIPTOR//PROTEIN SURFACE SHAPE |
| 3 | 0.0000218223 | LOOP PREDICTION//LOOP MODELING//STEPWISE FOLDING |
| 4 | 0.0000193154 | EMBL OUTSTN//PROT INFORMAT OURCE//HLTH SCI TECHNOL ICIR |
| 5 | 0.0000186413 | HP MODEL//STATISTICAL POTENTIALS//PROTEIN STRUCTURE PREDICTION |
| 6 | 0.0000185360 | EVOLUTIONARY BIOINFORMAT//CLOSED LOOPS//SKEW PAIRS |
| 7 | 0.0000164965 | MULTIPLE SEQUENCE ALIGNMENT//T COFFEE//STATISTICAL ALIGNMENT |
| 8 | 0.0000142901 | CORRELATED MUTATIONS//CONTACT PREDICTION//CONTACT MAP PREDICTION |
| 9 | 0.0000108302 | COMPUTATIONAL PROTEIN DESIGN//DEAD END ELIMINATION//COMPUTATIONAL ENZYME DESIGN |
| 10 | 0.0000084872 | BIOINFORMAT TELEMED//HYDROPHOBICITY DEFICIENCY//HELIX INTERACTIONS |