Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
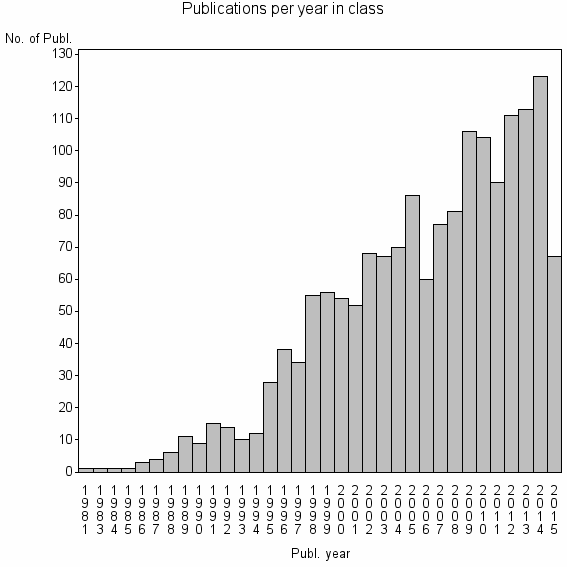
| 4962 | 1628 | 53.4 | 94% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | P TEFB | Author keyword | 123 | 74% | 6% | 91 |
| 2 | CDK9 | Author keyword | 61 | 66% | 4% | 57 |
| 3 | HEXIM1 | Author keyword | 56 | 89% | 2% | 25 |
| 4 | RNA POLYMERASE II | Author keyword | 34 | 19% | 10% | 157 |
| 5 | CYCLIN T1 | Author keyword | 30 | 68% | 2% | 26 |
| 6 | CYCLIN T | Author keyword | 26 | 87% | 1% | 13 |
| 7 | TRANSCRIPTION ELONGATION | Author keyword | 25 | 33% | 4% | 63 |
| 8 | DSIF | Author keyword | 22 | 68% | 1% | 19 |
| 9 | SPT5 | Author keyword | 15 | 68% | 1% | 13 |
| 10 | CTD PHOSPHATASE | Author keyword | 15 | 73% | 1% | 11 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | P TEFB | 123 | 74% | 6% | 91 | Search P+TEFB | Search P+TEFB |
| 2 | CDK9 | 61 | 66% | 4% | 57 | Search CDK9 | Search CDK9 |
| 3 | HEXIM1 | 56 | 89% | 2% | 25 | Search HEXIM1 | Search HEXIM1 |
| 4 | RNA POLYMERASE II | 34 | 19% | 10% | 157 | Search RNA+POLYMERASE+II | Search RNA+POLYMERASE+II |
| 5 | CYCLIN T1 | 30 | 68% | 2% | 26 | Search CYCLIN+T1 | Search CYCLIN+T1 |
| 6 | CYCLIN T | 26 | 87% | 1% | 13 | Search CYCLIN+T | Search CYCLIN+T |
| 7 | TRANSCRIPTION ELONGATION | 25 | 33% | 4% | 63 | Search TRANSCRIPTION+ELONGATION | Search TRANSCRIPTION+ELONGATION |
| 8 | DSIF | 22 | 68% | 1% | 19 | Search DSIF | Search DSIF |
| 9 | SPT5 | 15 | 68% | 1% | 13 | Search SPT5 | Search SPT5 |
| 10 | CTD PHOSPHATASE | 15 | 73% | 1% | 11 | Search CTD+PHOSPHATASE | Search CTD+PHOSPHATASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | P TEFB | 220 | 43% | 24% | 392 |
| 2 | ELONGATION FACTOR B | 144 | 80% | 5% | 89 |
| 3 | 7SK SNRNA | 94 | 75% | 4% | 68 |
| 4 | POSITIVE TRANSCRIPTION | 85 | 89% | 2% | 39 |
| 5 | LARGEST SUBUNIT | 81 | 40% | 10% | 160 |
| 6 | FACTOR P TEFB | 73 | 65% | 4% | 69 |
| 7 | CAPPING ENZYME | 54 | 38% | 7% | 112 |
| 8 | CARBOXYL TERMINAL DOMAIN | 52 | 29% | 9% | 151 |
| 9 | HIV 1 TAT | 51 | 24% | 11% | 185 |
| 10 | PROMONOCYTIC CELL LINES | 48 | 91% | 1% | 20 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Coupling mRNA processing with transcription in time and space | 2014 | 44 | 184 | 58% |
| Getting up to speed with transcription elongation by RNA polymerase II | 2015 | 5 | 132 | 57% |
| Controlling the elongation phase of transcription with P-TEFb | 2006 | 452 | 93 | 83% |
| Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans | 2012 | 164 | 97 | 57% |
| Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription | 2013 | 54 | 115 | 70% |
| Progression through the RNA Polymerase II CTD Cycle | 2009 | 286 | 45 | 67% |
| RNA Polymerase II Elongation Control | 2012 | 108 | 166 | 70% |
| Epigenetics in Alternative Pre-mRNA Splicing | 2011 | 253 | 91 | 52% |
| Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors | 2005 | 307 | 51 | 75% |
| The RNA polymerase II CTD coordinates transcription and RNA processing | 2012 | 98 | 253 | 62% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ROSALIND RUSSELL MED | 11 | 39% | 1.4% | 22 |
| 2 | REGULAT EXP S GENET | 10 | 73% | 0.5% | 8 |
| 3 | UMR 8541 | 4 | 21% | 1.2% | 19 |
| 4 | EXP S ENGN GRP | 4 | 42% | 0.5% | 8 |
| 5 | SICKLE CELL DIS | 4 | 13% | 1.7% | 27 |
| 6 | EDINBURGH SYST BIOL | 3 | 100% | 0.2% | 3 |
| 7 | GENET MOL GEN BIOL | 3 | 45% | 0.3% | 5 |
| 8 | GRP PHYS BIOCHEM | 3 | 50% | 0.2% | 4 |
| 9 | INSERM U685 | 2 | 67% | 0.1% | 2 |
| 10 | FISIOL BIOL MOL CELULARIFIBYNE CONICET | 2 | 50% | 0.2% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000159875 | MRNP BIOGENESIS METAB//DIS3//RRP6 |
| 2 | 0.0000128625 | MED12//MEDIATOR COMPLEX//MED19 |
| 3 | 0.0000126060 | TAT//TAT PROTEIN//TAR RNA |
| 4 | 0.0000125168 | POLYADENYLATION//ALTERNATIVE POLYADENYLATION//POLYA POLYMERASE |
| 5 | 0.0000110936 | TFIID//TFIIA//MOT1 |
| 6 | 0.0000103442 | NUT MIDLINE CARCINOMA//BROMODOMAIN//BRD4 |
| 7 | 0.0000094367 | P54NRB//POLYPYRIMIDINE TRACT BINDING PROTEIN ASSOCIATED SPLICING FACTOR//MATRIN 3 |
| 8 | 0.0000085070 | RPB7//RPB4//INTERSPECIFIC COMPLEMENTATION |
| 9 | 0.0000079082 | PIN1//ESS1//WW DOMAIN |
| 10 | 0.0000078990 | NUSG//TRANSCRIPTION TERMINATION//RHO FACTOR |