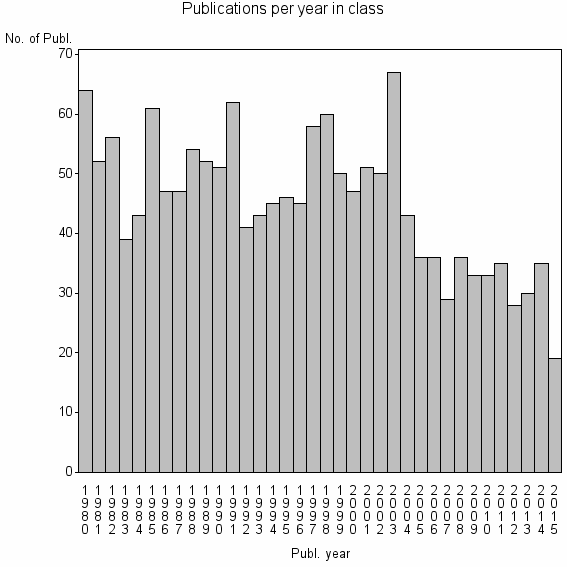
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 4978 | 1624 | 40.3 | 66% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 501 | 14604 | RNA POLYMERASE//SITE SPECIFIC RECOMBINATION//H NS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RNA POLYMERASE | Author keyword | 25 | 14% | 10% | 162 |
| 2 | ESCHERICHIA COLI RNA POLYMERASE | Author keyword | 23 | 72% | 1% | 18 |
| 3 | SIGMA70 | Author keyword | 17 | 68% | 1% | 15 |
| 4 | E COLI RNA POLYMERASE | Author keyword | 13 | 61% | 1% | 14 |
| 5 | GENE EXP S REGULAT SECT | Address | 13 | 71% | 1% | 10 |
| 6 | SIGMA SUBUNIT | Author keyword | 12 | 75% | 1% | 9 |
| 7 | PROKARYOTIC PROMOTER | Author keyword | 11 | 100% | 0% | 6 |
| 8 | UP ELEMENT | Author keyword | 9 | 55% | 1% | 11 |
| 9 | WAKSMAN MICROBIOL | Address | 6 | 18% | 2% | 32 |
| 10 | SIGMA FACTOR | Author keyword | 6 | 13% | 3% | 43 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA POLYMERASE | 25 | 14% | 10% | 162 | Search RNA+POLYMERASE | Search RNA+POLYMERASE |
| 2 | ESCHERICHIA COLI RNA POLYMERASE | 23 | 72% | 1% | 18 | Search ESCHERICHIA+COLI+RNA+POLYMERASE | Search ESCHERICHIA+COLI+RNA+POLYMERASE |
| 3 | SIGMA70 | 17 | 68% | 1% | 15 | Search SIGMA70 | Search SIGMA70 |
| 4 | E COLI RNA POLYMERASE | 13 | 61% | 1% | 14 | Search E+COLI+RNA+POLYMERASE | Search E+COLI+RNA+POLYMERASE |
| 5 | SIGMA SUBUNIT | 12 | 75% | 1% | 9 | Search SIGMA+SUBUNIT | Search SIGMA+SUBUNIT |
| 6 | PROKARYOTIC PROMOTER | 11 | 100% | 0% | 6 | Search PROKARYOTIC+PROMOTER | Search PROKARYOTIC+PROMOTER |
| 7 | UP ELEMENT | 9 | 55% | 1% | 11 | Search UP+ELEMENT | Search UP+ELEMENT |
| 8 | SIGMA FACTOR | 6 | 13% | 3% | 43 | Search SIGMA+FACTOR | Search SIGMA+FACTOR |
| 9 | SIGMA 70 | 5 | 54% | 0% | 7 | Search SIGMA+70 | Search SIGMA+70 |
| 10 | PROMOTER MELTING | 5 | 63% | 0% | 5 | Search PROMOTER+MELTING | Search PROMOTER+MELTING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NONTEMPLATE STRAND | 93 | 88% | 3% | 44 |
| 2 | SIGMA 70 SUBUNIT | 76 | 67% | 4% | 68 |
| 3 | LAC UV5 PROMOTER | 74 | 55% | 6% | 91 |
| 4 | OPEN COMPLEX FORMATION | 65 | 52% | 5% | 88 |
| 5 | COLI RNA POLYMERASE | 59 | 25% | 13% | 203 |
| 6 | SIGMA70 SUBUNIT | 54 | 65% | 3% | 51 |
| 7 | T4 ASIA | 48 | 100% | 1% | 17 |
| 8 | TRANSCRIPTION INITIATION | 39 | 19% | 11% | 186 |
| 9 | PROMOTER RECOGNITION | 28 | 36% | 4% | 64 |
| 10 | BACTERIOPHAGE T4 ASIA PROTEIN | 26 | 100% | 1% | 11 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Mechanism of Bacterial Transcription Initiation: RNA Polymerase - Promoter Binding, Isomerization to Initiation-Competent Open Complexes, and Initiation of RNA Synthesis | 2011 | 73 | 134 | 78% |
| Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective | 2014 | 13 | 107 | 81% |
| Advances in bacterial promoter recognition and its control by factors that do not bind DNA | 2008 | 122 | 144 | 50% |
| Bacterial RNA polymerases: the wholo story | 2003 | 239 | 55 | 65% |
| Protein family review - The sigma(70) family of sigma factors | 2003 | 193 | 23 | 74% |
| Anatomy of Escherichia coli sigma(70) promoters | 2007 | 70 | 102 | 41% |
| The regulation of bacterial transcription initiation | 2004 | 360 | 78 | 26% |
| Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners | 1999 | 108 | 98 | 69% |
| UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition | 2000 | 161 | 52 | 56% |
| Activating Transcription in Bacteria | 2012 | 48 | 137 | 31% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENE EXP S REGULAT SECT | 13 | 71% | 0.6% | 10 |
| 2 | WAKSMAN MICROBIOL | 6 | 18% | 2.0% | 32 |
| 3 | SYST MACROMOL SIGNALISAT | 2 | 67% | 0.1% | 2 |
| 4 | MOL GENET MICROORGANISMS | 2 | 26% | 0.4% | 7 |
| 5 | CNRS UMR 5236 | 1 | 50% | 0.1% | 2 |
| 6 | ENGN BIOTECHNOL HIGASHIMURA CHO | 1 | 100% | 0.1% | 2 |
| 7 | MOL GENET GENE BIOL | 1 | 40% | 0.1% | 2 |
| 8 | MOL GENET BACTERIA | 1 | 22% | 0.2% | 4 |
| 9 | UNITE PHYSICOCHIM MACROMOL BIOL | 1 | 15% | 0.3% | 5 |
| 10 | EA DOISY BIOCHEM MOL BIOL | 1 | 11% | 0.4% | 7 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000223131 | RPB7//RPB4//INTERSPECIFIC COMPLEMENTATION |
| 2 | 0.0000210966 | CYCLIC AMP RECEPTOR PROTEIN//CATABOLITE ACTIVATOR PROTEIN CAP//CAMP RECEPTOR PROTEIN |
| 3 | 0.0000205202 | PPGPP//STRINGENT RESPONSE//RPOS |
| 4 | 0.0000191294 | NUSG//TRANSCRIPTION TERMINATION//RHO FACTOR |
| 5 | 0.0000153309 | T7 RNA POLYMERASE//T7 LYSOZYME//PHAGE RNA POLYMERASE |
| 6 | 0.0000136962 | REX EXCLUSION//BACTERIOPHAGE LAMBDA LAMBDA//CI REXA REXB OPERON |
| 7 | 0.0000096899 | HRCA//RNA THERMOMETER//CIRCE |
| 8 | 0.0000084443 | TRP REPRESSOR//LAC REPRESSOR//LACTOSE REPRESSOR |
| 9 | 0.0000084058 | BACTERIOPHAGE T5//HOST PHAGE INTERACTION//LYTIC CONVERSION |
| 10 | 0.0000067498 | GENOME VECTOR//NIPPONKO KU//LONG ACCURATE PCR |