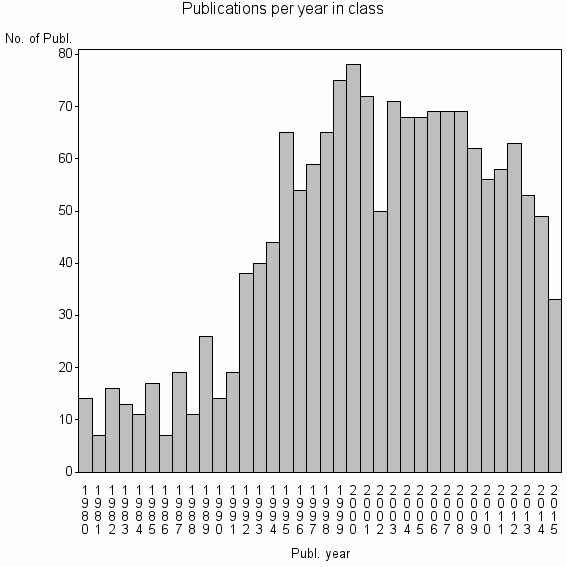
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 5115 | 1602 | 46.1 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 583 | 13522 | JOURNAL OF MAGNETIC RESONANCE//JOURNAL OF BIOMOLECULAR NMR//RESIDUAL DIPOLAR COUPLINGS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CROSS CORRELATED RELAXATION | Author keyword | 48 | 77% | 2% | 33 |
| 2 | N 15 RELAXATION | Author keyword | 29 | 51% | 3% | 41 |
| 3 | NMR SPIN RELAXATION | Author keyword | 24 | 82% | 1% | 14 |
| 4 | RELAXATION INTERFERENCE | Author keyword | 21 | 90% | 1% | 9 |
| 5 | CONFORMATIONAL EXCHANGE | Author keyword | 20 | 48% | 2% | 30 |
| 6 | PROTEIN DYNAMICS | Author keyword | 15 | 11% | 8% | 127 |
| 7 | SPECTRAL DENSITY MAPPING | Author keyword | 15 | 73% | 1% | 11 |
| 8 | BACKBONE DYNAMICS | Author keyword | 14 | 31% | 2% | 39 |
| 9 | RELAXATION DISPERSION | Author keyword | 13 | 39% | 2% | 26 |
| 10 | CHEMICAL EXCHANGE | Author keyword | 12 | 20% | 3% | 52 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CROSS CORRELATED RELAXATION | 48 | 77% | 2% | 33 | Search CROSS+CORRELATED+RELAXATION | Search CROSS+CORRELATED+RELAXATION |
| 2 | N 15 RELAXATION | 29 | 51% | 3% | 41 | Search N+15+RELAXATION | Search N+15+RELAXATION |
| 3 | NMR SPIN RELAXATION | 24 | 82% | 1% | 14 | Search NMR+SPIN+RELAXATION | Search NMR+SPIN+RELAXATION |
| 4 | RELAXATION INTERFERENCE | 21 | 90% | 1% | 9 | Search RELAXATION+INTERFERENCE | Search RELAXATION+INTERFERENCE |
| 5 | CONFORMATIONAL EXCHANGE | 20 | 48% | 2% | 30 | Search CONFORMATIONAL+EXCHANGE | Search CONFORMATIONAL+EXCHANGE |
| 6 | PROTEIN DYNAMICS | 15 | 11% | 8% | 127 | Search PROTEIN+DYNAMICS | Search PROTEIN+DYNAMICS |
| 7 | SPECTRAL DENSITY MAPPING | 15 | 73% | 1% | 11 | Search SPECTRAL+DENSITY+MAPPING | Search SPECTRAL+DENSITY+MAPPING |
| 8 | BACKBONE DYNAMICS | 14 | 31% | 2% | 39 | Search BACKBONE+DYNAMICS | Search BACKBONE+DYNAMICS |
| 9 | RELAXATION DISPERSION | 13 | 39% | 2% | 26 | Search RELAXATION+DISPERSION | Search RELAXATION+DISPERSION |
| 10 | CHEMICAL EXCHANGE | 12 | 20% | 3% | 52 | Search CHEMICAL+EXCHANGE | Search CHEMICAL+EXCHANGE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MODEL FREE APPROACH | 229 | 36% | 32% | 516 |
| 2 | BACKBONE DYNAMICS | 164 | 29% | 30% | 485 |
| 3 | MAGNETIC RESONANCE RELAXATION | 110 | 31% | 19% | 297 |
| 4 | SIDE CHAIN DYNAMICS | 101 | 61% | 7% | 109 |
| 5 | CHEMICAL SHIFT ANISOTROPY | 97 | 37% | 13% | 207 |
| 6 | TIME SCALE DYNAMICS | 84 | 65% | 5% | 79 |
| 7 | CAVITY MUTANT | 67 | 77% | 3% | 46 |
| 8 | DEUTERIUM SPIN PROBES | 58 | 83% | 2% | 33 |
| 9 | CHARACTERIZING CHEMICAL EXCHANGE | 52 | 76% | 2% | 37 |
| 10 | N 15 NMR RELAXATION | 39 | 29% | 7% | 114 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| NMR characterization of the dynamics of biomacromolecules | 2004 | 404 | 192 | 71% |
| Review - New tools provide new insights in NMR studies of protein dynamics | 2006 | 401 | 30 | 67% |
| Protein dynamics from NMR | 2000 | 326 | 63 | 75% |
| Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules | 2001 | 518 | 94 | 60% |
| NMR spectroscopy brings invisible protein states into focus | 2009 | 155 | 84 | 43% |
| Probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: An application to protein folding | 2008 | 97 | 32 | 56% |
| An introduction to NMR-based approaches for measuring protein dynamics | 2011 | 64 | 177 | 47% |
| Characterization of enzyme motions by solution NMR relaxation dispersion | 2008 | 75 | 49 | 61% |
| NMR probes of molecular dynamics: Overview and comparison with other techniques | 2001 | 247 | 148 | 76% |
| Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution | 2006 | 131 | 126 | 70% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | STRUCT MOL BIOL UNIT | 6 | 37% | 0.8% | 13 |
| 2 | PROT ENGN NETWORK EXCELLENCE | 6 | 15% | 2.3% | 37 |
| 3 | STRUCT BIOCOMP PROGRAMME | 3 | 60% | 0.2% | 3 |
| 4 | PROGRAM MOL STRUCT FUNCT | 2 | 10% | 1.3% | 21 |
| 5 | MOL GENET BIOCHEM CHEM | 2 | 67% | 0.1% | 2 |
| 6 | ONANCE MAGNET BIOMOL | 2 | 29% | 0.3% | 5 |
| 7 | MOL PROT SCI | 2 | 11% | 0.9% | 14 |
| 8 | BIO21 BIOTECHNOL MOL SCI | 1 | 50% | 0.1% | 2 |
| 9 | MED GENET CHEM | 1 | 100% | 0.1% | 2 |
| 10 | PROGRAM BIOPHYS MOL CELLULAR BIOL | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000229656 | RESIDUAL DIPOLAR COUPLINGS//RESIDUAL DIPOLAR COUPLING//ALIGNMENT TENSOR |
| 2 | 0.0000143295 | POLYPROLINE II//RANDOM COIL//COIL LIBRARY |
| 3 | 0.0000140529 | JOURNAL OF BIOMOLECULAR NMR//ULTRAFAST 2D NMR//CYANA |
| 4 | 0.0000130701 | 3J COUPLING CONSTANTS//DISTANCE GEOMETRY//TIME AVERAGED RESTRAINTS |
| 5 | 0.0000129941 | MODEL LUBRICANTS//DICYCLOHEXYL COMPOUNDS//OCTYL CHAINS |
| 6 | 0.0000093912 | RIBOSE AND NUCLEOBASE//ALTERNATE SITE SPECIFIC LABELING//DL323 |
| 7 | 0.0000082358 | ELASTIC NETWORK MODEL//ELASTIC NETWORK MODELS//NORMAL MODE ANALYSIS |
| 8 | 0.0000073290 | AFFINITY INDEX//H 1 SPIN LATTICE RELAXATION RATE//LIGAND MACROMOLECULES INTERACTION |
| 9 | 0.0000050046 | MAGIC ANGLE SPINNING//SOLID STATE NMR//DIPOLAR RECOUPLING |
| 10 | 0.0000048789 | FLUORESCENCE DEPOLARIZATION//TRANSPORT TRANSFORMAT BIOSYST//THERMODYNAM ETATS METASTABLES PHYS MOL |