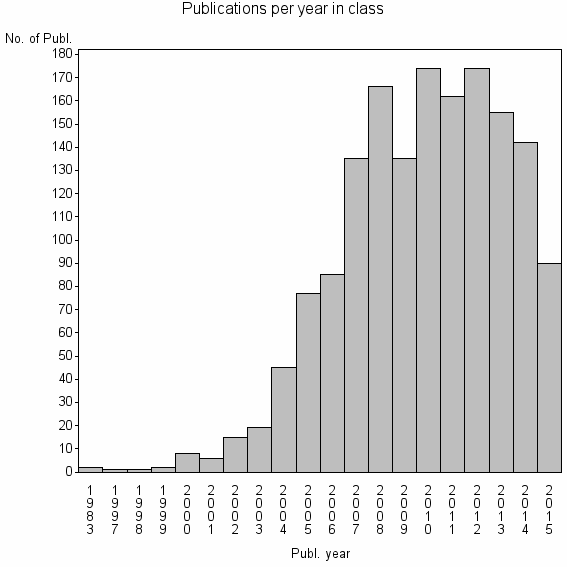
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 5163 | 1594 | 39.7 | 88% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1568 | 6715 | NEXT GENERATION SEQUENCING//RNA SEQ//COPY NUMBER VARIATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | COPY NUMBER VARIATION | Author keyword | 29 | 20% | 8% | 134 |
| 2 | CCL3L1 | Author keyword | 19 | 76% | 1% | 13 |
| 3 | PENNCNV | Author keyword | 13 | 80% | 1% | 8 |
| 4 | DNA COPY NUMBER | Author keyword | 10 | 37% | 1% | 22 |
| 5 | CRLMM | Author keyword | 6 | 80% | 0% | 4 |
| 6 | COPY NUMBER VARIATIONS | Author keyword | 6 | 20% | 2% | 25 |
| 7 | CCL3L | Author keyword | 6 | 100% | 0% | 4 |
| 8 | PARALOGUE RATIO TEST | Author keyword | 6 | 100% | 0% | 4 |
| 9 | COPY NUMBER VARIATION CNV | Author keyword | 5 | 27% | 1% | 16 |
| 10 | COPY NUMBER POLYMORPHISM | Author keyword | 4 | 36% | 1% | 9 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | COPY NUMBER VARIATION | 29 | 20% | 8% | 134 | Search COPY+NUMBER+VARIATION | Search COPY+NUMBER+VARIATION |
| 2 | CCL3L1 | 19 | 76% | 1% | 13 | Search CCL3L1 | Search CCL3L1 |
| 3 | PENNCNV | 13 | 80% | 1% | 8 | Search PENNCNV | Search PENNCNV |
| 4 | DNA COPY NUMBER | 10 | 37% | 1% | 22 | Search DNA+COPY+NUMBER | Search DNA+COPY+NUMBER |
| 5 | CRLMM | 6 | 80% | 0% | 4 | Search CRLMM | Search CRLMM |
| 6 | COPY NUMBER VARIATIONS | 6 | 20% | 2% | 25 | Search COPY+NUMBER+VARIATIONS | Search COPY+NUMBER+VARIATIONS |
| 7 | CCL3L | 6 | 100% | 0% | 4 | Search CCL3L | Search CCL3L |
| 8 | PARALOGUE RATIO TEST | 6 | 100% | 0% | 4 | Search PARALOGUE+RATIO+TEST | Search PARALOGUE+RATIO+TEST |
| 9 | COPY NUMBER VARIATION CNV | 5 | 27% | 1% | 16 | Search COPY+NUMBER+VARIATION+CNV | Search COPY+NUMBER+VARIATION+CNV |
| 10 | COPY NUMBER POLYMORPHISM | 4 | 36% | 1% | 9 | Search COPY+NUMBER+POLYMORPHISM | Search COPY+NUMBER+POLYMORPHISM |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ARRAY CGH DATA | 170 | 65% | 10% | 161 |
| 2 | STRUCTURAL VARIATION | 81 | 26% | 17% | 274 |
| 3 | CIRCULAR BINARY SEGMENTATION | 56 | 57% | 4% | 66 |
| 4 | SEGMENTAL DUPLICATIONS | 48 | 25% | 10% | 165 |
| 5 | SNP GENOTYPING DATA | 46 | 50% | 4% | 65 |
| 6 | CGH DATA | 44 | 59% | 3% | 50 |
| 7 | GENOTYPING DATA | 33 | 75% | 2% | 24 |
| 8 | COPY NUMBER VARIATION | 30 | 13% | 13% | 215 |
| 9 | VARIANT ASSAYS | 24 | 91% | 1% | 10 |
| 10 | CCL3L1 | 22 | 66% | 1% | 21 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| A copy number variation map of the human genome | 2015 | 3 | 90 | 39% |
| Copy number polymorphism in plant genomes | 2014 | 14 | 91 | 29% |
| Correlating Multiallelic Copy Number Polymorphisms with Disease Susceptibility | 2013 | 18 | 105 | 51% |
| Copy Number Variation in Human Health, Disease, and Evolution | 2009 | 328 | 166 | 23% |
| Human Copy Number Variation and Complex Genetic Disease | 2011 | 75 | 130 | 31% |
| Impacts of Variation in the Human Genome on Gene Regulation | 2013 | 23 | 45 | 33% |
| Mechanisms of change in gene copy number | 2009 | 362 | 145 | 17% |
| Copy number variation in the genomes of domestic animals | 2012 | 28 | 90 | 48% |
| Phenotypic impact of genomic structural variation: insights from and for human disease | 2013 | 47 | 145 | 16% |
| Structural variation in the human genome | 2006 | 750 | 87 | 32% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SHANDONG PROV ANIM DIS CONTROL BREEDING | 4 | 46% | 0.4% | 6 |
| 2 | HLTH SOLUT GRP | 3 | 43% | 0.4% | 6 |
| 3 | INTEGRATED GENOME POLYMORPHISM | 3 | 25% | 0.7% | 11 |
| 4 | GENET GENOM ANAL | 3 | 60% | 0.2% | 3 |
| 5 | JNU HKUST JOINT | 3 | 32% | 0.4% | 7 |
| 6 | GRP MOL GENET SYST BIOL | 2 | 67% | 0.1% | 2 |
| 7 | JMP GENOM | 2 | 67% | 0.1% | 2 |
| 8 | BREAST CANC FUNCT GEN | 1 | 100% | 0.1% | 2 |
| 9 | COMMUNITY ENGAGED TRANSLAT | 1 | 50% | 0.1% | 2 |
| 10 | EPIDEMIOL PUBL HLTH MD3 | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000180362 | SUBTELOMERIC REARRANGEMENTS//IDIOPATHIC MENTAL RETARDATION//MBD5 |
| 2 | 0.0000146051 | ZNF804A//MCLAUGHLIN//MOL CLIN NEUROBIOL |
| 3 | 0.0000104045 | INTER SIMPLE SEQUENCE REPEAT PCR//BIOPHYS CYTOMETRY//HOSP UNIV SALAMANCA IBSAL |
| 4 | 0.0000102895 | GENOME ASSEMBLY//VARIANT CALLING//TARGET ENRICHMENT |
| 5 | 0.0000088993 | EVOLUT CANC//PETOS PARADOX//TRANSLAT CANC THER EUT |
| 6 | 0.0000081933 | COMPLEX CHROMOSOMAL REARRANGEMENT//COMPLEX CHROMOSOME REARRANGEMENT CCR//COMPLEX CHROMOSOMAL REARRANGEMENTS |
| 7 | 0.0000065754 | ZNF703//C8ORF4//8P12 |
| 8 | 0.0000061830 | WHOLE GENOME AMPLIFICATION//MULTIPLE DISPLACEMENT AMPLIFICATION//PHI29 DNA POLYMERASE |
| 9 | 0.0000061500 | SMITH MAGENIS SYNDROME//RAI1//POTOCKI LUPSKI SYNDROME |
| 10 | 0.0000055039 | CLIN BREAST CARE PROJECT//16Q243//CHROMOSOMAL SCANNING |